Phase and context shape the function of composite oncogenic mutations
Phase and context shape the function of composite oncogenic mutations"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Cancers develop as a result of driver mutations1,2 that lead to clonal outgrowth and the evolution of disease3,4. The discovery and functional characterization of individual driver
mutations are central aims of cancer research, and have elucidated myriad phenotypes5 and therapeutic vulnerabilities6. However, the serial genetic evolution of mutant cancer genes7,8 and
the allelic context in which they arise is poorly understood in both common and rare cancer genes and tumour types. Here we find that nearly one in four human tumours contains a composite
mutation of a cancer-associated gene, defined as two or more nonsynonymous somatic mutations in the same gene and tumour. Composite mutations are enriched in specific genes, have an elevated
rate of use of less-common hotspot mutations acquired in a chronology driven in part by oncogenic fitness, and arise in an allelic configuration that reflects context-specific selective
pressures. _cis_-acting composite mutations are hypermorphic in some genes in which dosage effects predominate (such as _TERT_), whereas they lead to selection of function in other genes
(such as _TP53_). Collectively, composite mutations are driver alterations that arise from context- and allele-specific selective pressures that are dependent in part on gene and mutation
function, and which lead to complex—often neomorphic—functions of biological and therapeutic importance. Access through your institution Buy or subscribe This is a preview of subscription
content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access
subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article
* Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn
about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MUTATIONAL LANDSCAPE OF CANCER-DRIVER GENES ACROSS HUMAN CANCERS Article
Open access 07 August 2023 A COMPENDIUM OF MUTATIONAL CANCER DRIVER GENES Article 10 August 2020 HIGHER ORDER GENETIC INTERACTIONS SWITCH CANCER GENES FROM TWO-HIT TO ONE-HIT DRIVERS Article
Open access 03 December 2021 DATA AVAILABILITY All mutational data from the prospective sequencing cohort are available at http://download.cbioportal.org/composite_mutations_maf.txt.gz.
Mutational data from The Cancer Genome Atlas were acquired from https://gdc.cancer.gov/about-data/publications/pancanatlas. RNA sequencing data have been deposited in the Gene Expression
Omnibus with accession number GSE136295. All other genomic and clinical data accompany the Article, and are available in the Extended Data and Supplementary Information. All other materials
are available upon request from the corresponding authors. CODE AVAILABILITY Source code for these analyses is available at https://github.com/taylor-lab/composite-mutations. REFERENCES *
Vogelstein, B. et al. Cancer genome landscapes. _Science_ 339, 1546–1558 (2013). ADS CAS PubMed PubMed Central Google Scholar * Garraway, L. A. & Lander, E. S. Lessons from the
cancer genome. _Cell_ 153, 17–37 (2013). CAS PubMed Google Scholar * Cairns, J. Mutation selection and the natural history of cancer. _Nature_ 255, 197–200 (1975). ADS CAS PubMed
Google Scholar * Nowell, P. C. The clonal evolution of tumor cell populations. _Science_ 194, 23–28 (1976). ADS CAS PubMed Google Scholar * Hanahan, D. & Weinberg, R. A. Hallmarks
of cancer: the next generation. _Cell_ 144, 646–674 (2011). Article CAS PubMed Google Scholar * Hyman, D. M., Taylor, B. S. & Baselga, J. Implementing genome-driven oncology. _Cell_
168, 584–599 (2017). CAS PubMed PubMed Central Google Scholar * Knudson, A. G., Jr. Mutation and cancer: statistical study of retinoblastoma. _Proc. Natl Acad. Sci. USA_ 68, 820–823
(1971). ADS PubMed PubMed Central Google Scholar * Bielski, C. M. et al. Widespread selection for oncogenic mutant allele imbalance in cancer. _Cancer Cell_ 34, 852–862.e4 (2018). CAS
PubMed PubMed Central Google Scholar * Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. _Nature_ 499, 214–218 (2013). ADS CAS
PubMed PubMed Central Google Scholar * Jin, G. et al. Disruption of wild-type IDH1 suppresses d-2-hydroxyglutarate production in IDH1-mutated gliomas. _Cancer Res_. 73, 496–501 (2013).
CAS PubMed Google Scholar * Mueller, S. et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. _Nature_ 554, 62–68 (2018). ADS CAS PubMed PubMed Central
Google Scholar * Chang, M. T. et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. _Nat. Biotechnol_. 34, 155–163 (2016). CAS
PubMed Google Scholar * Chang, M. T. et al. Accelerating discovery of functional mutant alleles in cancer. _Cancer Discov_. 8, 174–183 (2018). CAS PubMed Google Scholar * Intlekofer, A.
M. et al. Acquired resistance to IDH inhibition through _trans_ or _cis_ dimer-interface mutations. _Nature_ 559, 125–129 (2018). ADS CAS PubMed PubMed Central Google Scholar * Hidaka,
N. et al. Most T790M mutations are present on the same _EGFR_ allele as activating mutations in patients with non-small cell lung cancer. _Lung Cancer_ 108, 75–82 (2017). PubMed Google
Scholar * Gainor, J. F. et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. _Cancer Discov_. 6, 1118–1133 (2016). CAS
PubMed PubMed Central Google Scholar * Kobayashi, S. et al. _EGFR_ mutation and resistance of non-small-cell lung cancer to gefitinib. _N. Engl. J. Med_. 352, 786–792 (2005). CAS PubMed
Google Scholar * Vasan, N. et al. Double _PIK3CA_ mutations _in cis_ increase oncogenicity and sensitivity to PI3Kα inhibitors. _Science_ 366, 714–723 (2019). ADS CAS PubMed PubMed
Central Google Scholar * Chen, Z. et al. _EGFR_ somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations:
one-third of doublets occur at five pairs of amino acids. _Oncogene_ 27, 4336–4343 (2008). CAS PubMed Google Scholar * Huang, F. W. et al. Highly recurrent _TERT_ promoter mutations in
human melanoma. _Science_ 339, 957–959 (2013). ADS CAS PubMed PubMed Central Google Scholar * Bell, R. J. A. et al. The transcription factor GABP selectively binds and activates the
mutant TERT promoter in cancer. _Science_ 348, 1036–1039 (2015). ADS CAS PubMed PubMed Central Google Scholar * Berenjeno, I. M. et al. Oncogenic _PIK3CA_ induces centrosome
amplification and tolerance to genome doubling. _Nat. Commun_. 8, 1773 (2017). ADS PubMed PubMed Central Google Scholar * Kinross, K. M. et al. An activating _Pik3ca_ mutation coupled
with _Pten_ loss is sufficient to initiate ovarian tumorigenesis in mice. _J. Clin. Invest_. 122, 553–557 (2012). CAS PubMed PubMed Central Google Scholar * Madsen, R. R. et al.
Oncogenic _PIK3CA_ promotes cellular stemness in an allele dose-dependent manner. _Proc. Natl Acad. Sci. USA_ 116, 8380–8389 (2019). CAS PubMed PubMed Central Google Scholar * Hyman, D.
M. et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. _Drug Discov. Today_ 20,
1422–1428 (2015). CAS PubMed PubMed Central Google Scholar * Cheng, D. T. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a
hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. _J. Mol. Diagn_. 17, 251–264 (2015). CAS PubMed PubMed Central Google Scholar *
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. _Nat. Med_. 23, 703–713 (2017). CAS PubMed PubMed Central
Google Scholar * Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. _JCO Precis. Oncol_. 1, 1–16 (2017). Google Scholar * Campbell, B. B. et al. Comprehensive analysis of
hypermutation in human cancer. _Cell_ 171, 1042–1056.e10 (2017). CAS PubMed PubMed Central Google Scholar * Niu, B. et al. MSIsensor: microsatellite instability detection using paired
tumor-normal sequence data. _Bioinformatics_ 30, 1015–1016 (2014). CAS PubMed Google Scholar * Middha, S. et al. Reliable pan-cancer microsatellite instability assessment by using
targeted next-generation sequencing data. _JCO Precis. Oncol_. 1, 1–17 (2017). Google Scholar * Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Campbell, P. J. & Stratton, M. R.
Deciphering signatures of mutational processes operative in human cancer. _Cell Rep_. 3, 246–259 (2013). CAS PubMed PubMed Central Google Scholar * Dixon, P. VEGAN, a package of R
functions for community ecology. _J. Veg. Sci_. 14, 927–930 (2003). Google Scholar * Smedley, D. et al. The BioMart community portal: an innovative alternative to large, centralized data
repositories. _Nucleic Acids Res_. 43, W589–W598 (2015). CAS PubMed PubMed Central Google Scholar * Forbes, S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in
human cancer. _Nucleic Acids Res_. 43, D805–D811 (2015). CAS PubMed Google Scholar * Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. _Nature_ 500, 415–421
(2013). CAS PubMed PubMed Central Google Scholar * Alexandrov, L. et al. The repertoire of mutational signatures in human cancer. _Nature_ 578, 94–101 (2020). ADS CAS PubMed PubMed
Central Google Scholar * Pich, O. et al. Somatic and germline mutation periodicity follow the orientation of the DNA minor groove around nucleosomes. _Cell_ 175, 1074–1087.e18 (2018). CAS
PubMed Google Scholar * Sabarinathan, R., Mularoni, L., Deu-Pons, J., Gonzalez-Perez, A. & López-Bigas, N. Nucleotide excision repair is impaired by binding of transcription factors
to DNA. _Nature_ 532, 264–267 (2016). ADS CAS PubMed Google Scholar * Buisson, R. et al. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features.
_Science_ 364, eaaw2872 (2019). ADS CAS PubMed PubMed Central Google Scholar * Hess, J. M. et al. Passenger hotspot mutations in cancer. _Cancer Cell_ 36, 288–301.e14 (2019). CAS
PubMed PubMed Central Google Scholar * Needleman, S. B. & Wunsch, C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. _J. Mol.
Biol_. 48, 443–453 (1970). CAS PubMed Google Scholar * McGranahan, N. et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. _Sci.
Transl. Med_. 7, 283ra54 (2015). PubMed PubMed Central Google Scholar * Dimitrova, N. et al. Stromal expression of miR-143/145 promotes neoangiogenesis in lung cancer development. _Cancer
Discov_. 6, 188–201 (2016). CAS PubMed Google Scholar * Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. _Bioinformatics_ 30,
2114–2120 (2014). CAS PubMed PubMed Central Google Scholar * Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. _Bioinformatics_ 29, 15–21 (2013). CAS PubMed Google Scholar *
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. _Bioinformatics_ 30, 923–930 (2014). CAS PubMed
Google Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. _Genome Biol_. 15, 550 (2014). PubMed PubMed
Central Google Scholar * Bult, C. J., Blake, J. A., Smith, C. L., Kadin, J. A. & Richardson, J. E. Mouse genome database (MGD) 2019. _Nucleic Acids Res_. 47, D801–D806 (2019). CAS
PubMed Google Scholar * Khan, A. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. _Nucleic Acids Res_. 46, D260–D266
(2018). CAS PubMed Google Scholar * Tan, G. & Lenhard, B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. _Bioinformatics_ 32, 1555–1556 (2016).
CAS PubMed PubMed Central Google Scholar * Touzet, H. & Varré, J.-S. Efficient and accurate _P_-value computation for position weight matrices. _Algorithms Mol. Biol_. 2, 15 (2007).
PubMed PubMed Central Google Scholar * Supek, F. & Lehner, B. Clustered mutation signatures reveal that error-prone DNA repair targets mutations to active genes. _Cell_ 170,
534–547.e23 (2017). CAS PubMed Google Scholar * Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. _Cell_ 149, 979–993 (2012). CAS PubMed PubMed
Central Google Scholar Download references ACKNOWLEDGEMENTS We thank the members of the E.R. and B.S.T. laboratories for discussion and support. This work was supported by National
Institutes of Health awards P30 CA008748, P01 CA087497 (S.W.L.), U54 OD020355 (S.W.L. and B.S.T.), R01 CA207244 (B.S.T.), R01 CA204749 (B.S.T.), R01 CA245069 (B.S.T.); Brown Performance
Group ICI Fund (N.V. and E.R.), Society of MSK (N.V. and E.R.), American Cancer Society, Anna Fuller Fund and the Josie Robertson Foundation (B.S.T.). F.J.S.-R. is an HHMI Hanna Gray Fellow
supported in part by an MSKCC Translational Research Oncology Training Fellowship (T32-CA160001). S.W.L. is an investigator of the Howard Hughes Medical Institute. AUTHOR INFORMATION AUTHORS
AND AFFILIATIONS * Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA Alexander N. Gorelick, Craig M. Bielski, Neil Vasan, Alexander V.
Penson, Noah D. Friedman & Barry S. Taylor * Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY, USA Alexander N. Gorelick, Craig M.
Bielski, Evan Biederstedt, Alexander V. Penson, Noah D. Friedman, Nikolaus Schultz, Ed Reznik & Barry S. Taylor * Cancer Biology and Genetics Program, Memorial Sloan Kettering Cancer
Center, New York, NY, USA Francisco J. Sánchez-Rivera, Yu-Jui Ho, Timour Baslan & Scott W. Lowe * Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, NY, USA
Yanyan Cai & Maurizio Scaltriti * Marie-Josee and Henry R. Kravis Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York, NY, USA Philip Jonsson, Allison L.
Richards, Chaitanya Bandlamudi, Nikolaus Schultz, Ed Reznik & Barry S. Taylor * Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA Neil Vasan * Weill
Cornell Medical College, New York, NY, USA Nikolaus Schultz & Barry S. Taylor * Howard Hughes Medical Institute, New York, NY, USA Scott W. Lowe Authors * Alexander N. Gorelick View
author publications You can also search for this author inPubMed Google Scholar * Francisco J. Sánchez-Rivera View author publications You can also search for this author inPubMed Google
Scholar * Yanyan Cai View author publications You can also search for this author inPubMed Google Scholar * Craig M. Bielski View author publications You can also search for this author
inPubMed Google Scholar * Evan Biederstedt View author publications You can also search for this author inPubMed Google Scholar * Philip Jonsson View author publications You can also search
for this author inPubMed Google Scholar * Allison L. Richards View author publications You can also search for this author inPubMed Google Scholar * Neil Vasan View author publications You
can also search for this author inPubMed Google Scholar * Alexander V. Penson View author publications You can also search for this author inPubMed Google Scholar * Noah D. Friedman View
author publications You can also search for this author inPubMed Google Scholar * Yu-Jui Ho View author publications You can also search for this author inPubMed Google Scholar * Timour
Baslan View author publications You can also search for this author inPubMed Google Scholar * Chaitanya Bandlamudi View author publications You can also search for this author inPubMed
Google Scholar * Maurizio Scaltriti View author publications You can also search for this author inPubMed Google Scholar * Nikolaus Schultz View author publications You can also search for
this author inPubMed Google Scholar * Scott W. Lowe View author publications You can also search for this author inPubMed Google Scholar * Ed Reznik View author publications You can also
search for this author inPubMed Google Scholar * Barry S. Taylor View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS A.N.G., E.R. and B.S.T.
conceived the study. C.M.B., E.B., P.J., A.V.P., A.L.R., N.D.F., C.B., N.S., E.R. and B.S.T. assisted with genomic data collection and analytical methodology development. F.J.S.-R., Y.C.,
N.V., M.S. and S.W.L. designed and performed the experiments. Y.J.H. and T.B. assisted with RNA sequencing. A.N.G., E.R. and B.S.T. wrote the manuscript with input from all authors.
CORRESPONDING AUTHORS Correspondence to Ed Reznik or Barry S. Taylor. ETHICS DECLARATIONS COMPETING INTERESTS N.V. reports advisory board activities for Novartis and consulting activities
for Petra Pharmaceuticals. M.S. has received research funding from Puma Biotechnology, Daiichi-Sankio, Immunomedics, Targimmune and Menarini Ricerche; is a cofounder of Medendi.org, and is
on the advisory boards of the Bioscience Institute and Menarini Ricerche. S.W.L. is a founder and scientific advisory board member of Oric Pharmaceuticals, Mirimus, Inc. and Blueprint
Medicines; and is on the scientific advisory boards of Constellation Pharmaceuticals, Petra Pharmaceuticals and PMV Pharmaceuticals. B.S.T. reports receiving honoria and research funding
from Genentech and Illumina, and advisory board activities for Boehringer Ingelheim and Loxo Oncology, a wholly owned subsidiary of Eli Lilly, Inc. All stated activities were outside of the
work described here. The other authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature_ thanks Moritz Gerstung, Mark Lackner and the other, anonymous,
reviewer(s) for their contribution to the peer review of this work. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
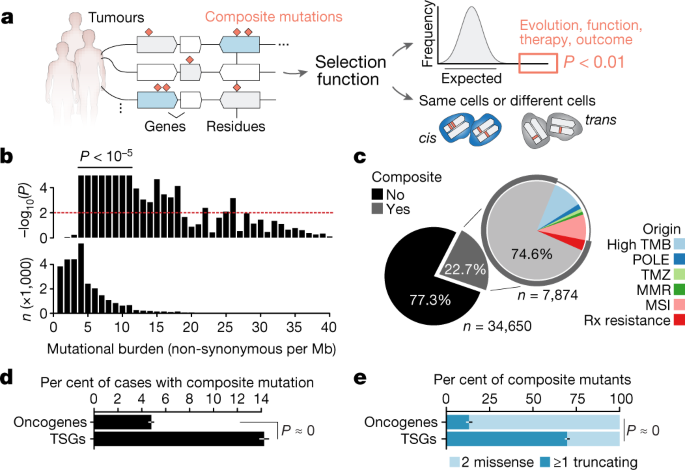
affiliations. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIG. 1 STUDY COHORT AND RATES OF COMPOSITE MUTATIONS. A, Distribution of cancer types in the study cohort. B, The rate of
composite mutations (22.7% of all tumours) compared to a simulated background rate (black, _P_ = 10−5 from one-sided permutation test for enrichment with 100,000 random permutation-based
simulations (no permutation exceeded observed value)). C, The observed rate of composite mutations in the primary untreated cancers of the TCGA cohort (_n_ = 10,908 solid tumours) when
controlling for gene content for consistency with the targeted sequencing panel of the prospective cohort studied here. The null distribution from sampling (Methods) is shown in black. D,
The observed and expected rate of composite mutations in tumours of the indicated tumour mutational burden (as in Fig. 1b, _n_ = 30,505 biologically independent tumour samples with tumour
mutational burden ≤ 40, _P_ = 1 × 10−9 from two-sided Wilcoxon signed-rank test). EXTENDED DATA FIG. 2 SOURCES OF LOCAL HYPERMUTATION. A, The number of composite mutations comprising two or
more constituent variants (top) and the distribution of likely causative mutational signatures among them (bottom). Composite mutants comprising greater than three mutations were
increasingly produced by APOBEC-associated mutagenesis, indicative of localized hypermutation53,54, but accounted for a minority of events cohort-wide. B, Left, the somatic mutational data
in the study cohort reflect the elevated mutation rates previously observed at both the positions closest to the nucleosome dyad as well as DNA bound to active transcription-factor binding
sites38,39. However, mutations arising in composite events were proportionally less often proximal to such sites (defined here as within the full width at half maximum of the peak of
mutation rate (red)) than were singleton mutations (right, _P_ = 10−27 and 10−47, respectively; two-sided two-sample _Z_-test, _n_ = 323,883 single-nucleotide substitutions arising in 471
biologically distinct melanoma samples). EXTENDED DATA FIG. 3 NUMBER AND DISTRIBUTION OF COMPOSITE EVENTS ACROSS GENES. A, The number and percentage of cases in the study cohort containing
composite mutations in the indicated genes (right) juxtaposed to their overall mutation rate (left). Genes with a significant enrichment of composite mutations are shown (_Q_ < 0.01,
FDR-adjusted _P_ values from one-sided binomial test for enrichment, _n_ = 26,997 as in Fig. 2b), limited to the top 10 genes by significance in each category of gene function, unless fewer.
B, The significance of enrichment for composite mutations (_n_ and statistical tests as described in A and Fig. 2b) limited to 168 oncogenes. EXTENDED DATA FIG. 4 _CIS_ COMPOSITE
SECONDARY-RESISTANCE MUTATIONS. The _cis_ composite mutations classified as arising in post-treatment specimens due to acquired resistance to one of several molecularly targeted therapies in
the study cohort. EXTENDED DATA FIG. 5 PHENOTYPIC CHARACTERIZATION OF _TP53_ COMPOSITE MUTANTS. A, _TP53__R280T/E287D_ mutant lung adenocarcinoma. Left, mutant allele fractions of clonal
_TP53_ mutations consistent with loss of wild-type _TP53_ (error bars, 95% binomial confidence intervals). Expected mutant allele fractions of different copy number states are shown as
horizontal lines. Mutant _KEAP1_ in the same tumour (with LOH) is shown for reference. Right, spanning reads indicating _cis_ mutations. B, Right and left, _Trp53_ and _Cdkn1a_ mRNA
expression in _Kras__G12D/+__Trp53__Mut_ mouse lung cancer cells expressing distinct _Trp53_ genotypes. Bars, average of three replicates, error bars are 95% confidence intervals. C, The
aggregate _Z_-score per replicate for the mRNA expression of canonical p53-target genes (_n_ = 3 replicates per allele; box centre is median, edges are 25% and 75% quartiles, whiskers are
minimum and maximum of the most extreme values). D, Principal component analysis of the transcriptomes of _Trp53_ genotypes (_n_ = 3 replicates shown per condition). E, Dendrogram as in Fig.
3f, indicating the genes of interest (effectors of the AP-1 transcription factor network (PID_AP1_PATHWAY; _Q_ = 1.4 × 10−7 based on computed overlap (using mSigDB) with _n_ = 5,501 gene
sets from the curated C2 collection)). F, The prevalence of _TP53__R280T_ and _TP53__E287D_ mutations (top), and the fraction arising as composite mutants (bottom). The corresponding mouse
alleles are given in parentheses. G, Principal component analysis of the transcriptomes of the _Trp53__R277K/E282K_ composite mutation genotypes (as in D). _n_ = 3 replicates per allele. H,
The percentage of GFP+ FACS-purified _Kras__G12D/+__Trp53__−/−_ lung adenocarcinoma cells stably transduced with pMIG-empty or pMIG-p53-R277T-E284D, and cultured in vitro for 10 days in a
60:40 mixture with untransduced parental cells. Bar indicates mean, error bars are s.d., _n_ = 3 independent infections. I, Overall survival of immunocompromised mice bearing lung tumours of
the indicated _Trp53_ genotypes generated by tail vein injection of stably transduced and FACS-purified _Kras__G12D/+__Trp53__−/−_ lung adenocarcinoma cells (_n_ = 100,000 cells). EXTENDED
DATA FIG. 6 SATURATION ANALYSIS OF GENES FOR COMPOSITE MUTATION DETECTION. Down-sampling indicates the number of residues identified as enriched for arising in composite mutations in each of
four genes (_Q_ < 0.1, FDR-adjusted one-sided Fisher’s exact tests as in Fig. 4a; _n_ = 1,000–26,997 patients per down-sample) as a function of the number of tumours sequenced (LOESS fit
is shown with 95% confidence interval). Four genes that accounted for the greatest proportion of all enriched residues detected are shown (Fig. 4a). _EGFR_ appears to reach saturation for
discovery of residues enriched for arising in composite, whereas the other genes have not yet reached saturation for discovery at the current cohort size. EXTENDED DATA FIG. 7 MUTATIONAL
SIGNATURE ATTRIBUTION AMONG COMPOSITE MUTATIONS. A, The fraction of all composite mutations identified here in which one or both individual mutations could be unambiguously attributed to an
established mutational signature. The majority of composite variants could not be directly attributed to APOBEC, ultraviolet, smoking or other known mutational signatures. B, The fraction of
composite mutations per gene in which one or both variants could be attributed to an established mutational signature. EXTENDED DATA FIG. 8 CONDITIONAL MUTANT ALLELES. A, The number of
affected cases containing each of the indicated somatic mutations in _TERT_, _EGFR_ or _PIK3CA_ as either individual mutations (top) or as part of composite mutants (bottom). Conditional
mutations were defined as those statistically enriched for arising as part of composite mutations, but seldom as individual hotspot mutations in cancer (predominantly accompanied by a second
somatic mutation). B, The incidence of _TERT_ promoter mutations and the fraction arising as composite mutations (orange). Bottom, the co-occurrence and mutual exclusivity of composite
mutations in the _TERT_ promoter (The _P_ values for _n_ = 5 and 6 co-occurring mutations are 0.002 and 3 × 10−7, respectively, and for 0 mutually exclusive mutations is 1 × 10−25; two-sided
Fisher’s exact test, _n_ = 29,507 patients). C, Transcription factor GABPA binding affinity for mutant and wild-type _TERT_ promoter sequences at the 228G>A, 250G>A and the
conditional 205G>A allele. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION This file contains a guide for Supplementary Tables 1-5. REPORTING SUMMARY SUPPLEMENTARY TABLES This file
contains Supplementary Tables 1-5 – see Supplementary Information document for full guide. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gorelick,
A.N., Sánchez-Rivera, F.J., Cai, Y. _et al._ Phase and context shape the function of composite oncogenic mutations. _Nature_ 582, 100–103 (2020). https://doi.org/10.1038/s41586-020-2315-8
Download citation * Received: 07 September 2019 * Accepted: 06 April 2020 * Published: 27 May 2020 * Issue Date: 04 June 2020 * DOI: https://doi.org/10.1038/s41586-020-2315-8 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
'as a black man, i have double the risk of prostate cancer'Apprentice star Tim Campbell has spoken out about how as a Black man he has "double the risk" of prostate canc...
How long does it take to plan a wedding? The ultimate timeline and checklistAbigail Buchanan 14 June 2021 9:58am BST It’s been said the course of love never did run smooth, and wedding planning is...
Hidden hearing loss and damageIf you think your hearing is fine, a new report from the Centers for Disease Control and Prevention (CDC) should make yo...
Supporting transitioning trans people is not conversion therapy. It is the right thing to do. | thearticleIn a recent article for this site, philosopher Dr. Kathleen Stock expressed concern over the definition of conversion th...
Roger scruton is dead, but his books and ideas will live on | thearticleSir Roger Scruton is dead. The knight errant of modern philosophy, the Edmund Burke of our time, the pimpernel of dissid...
Latests News
Phase and context shape the function of composite oncogenic mutationsABSTRACT Cancers develop as a result of driver mutations1,2 that lead to clonal outgrowth and the evolution of disease3,...
Balloon row a ‘surface wound’ for us-china ties: singapore’s george yeoFormer Singapore foreign minister George Yeo said the current US-China tensions over the shootdown of a Chinese balloon ...
Spring is coming in europe. When will britain emerge from lockdown? | thearticleAcross Europe, spring is here. Little by little, life is returning to a continent that has been in suspended animation s...
The cost of lies | TheArticleWhen Kim Philby defected to the USSR in 1963, he got a surprise. Even though he’d spent nearly thirty years as the KGB’s...
TINA for People with CancerMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...