Engineered trimeric ace2 binds viral spike protein and locks it in “three-up” conformation to potently inhibit sars-cov-2 infection
Engineered trimeric ace2 binds viral spike protein and locks it in “three-up” conformation to potently inhibit sars-cov-2 infection"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Dear Editor, Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a severe global pandemic. Following SARS-CoV,
SARS-CoV-2 is yet another emergent beta-coronavirus threatening human health.1 However, seventeen years after the SARS pandemic, no targeted vaccines or therapeutics have been approved for
SARS, while some of them might have held promise for treating COVID-19. Many neutralizing antibodies against SARS-CoV-2 are currently being developed. However, RNA viruses are known to have
high mutation rates. Many SARS-CoV-2 mutations have already been identified such as D614G,2 and these resultant mutation strains might escape the effects of the current SARS-CoV-2
neutralizing antibodies. The appearance of COVID-19 after SARS indicates the likely emergence of other coronavirus pandemics in the future. Thus, therapeutics broadly effective against
SARS-CoV-2 and mutants, even other SARS-CoV-2-related coronaviruses (SARSr-CoVs), are highly desirable. Both SARS-CoV-2 and SARS-CoV bind ACE2 for cell entry, suggesting a general use of
ACE2 by SARS-CoV-2 mutants and future related coronaviruses. Therefore, proteins engineered based on wild-type ACE2 might exhibit the best broad neutralizing activity and avoid the
mutational escape. Coronavirus infection can cause shedding of ACE2 resulting in a decreased level of ACE2 expression.3,4 This phenomenon is closely related to acute lung injury, and
replenishing soluble ACE2 could alleviate acute respiratory distress syndrome (ARDS).3,4 It has been shown that recombinant soluble ACE2 could inhibit SARS-CoVs infection in cells and
organoids.5,6 One clinical trial (NCT04335136) was also registered to use recombinant ACE2 to treat COVID-19. However, recombinant soluble ACE2 inhibits viral infection at relatively high
concentrations,5,6 therefore, it may not be an optimal inhibitor. Since spike proteins of SARS-CoVs function as trimers,7 we reasoned that an engineered trimeric ACE2 protein could
potentially bind up to three receptor binding domains (RBD) on the spike protein, which would dramatically increase binding affinity and thus potently inhibit SARS-CoVs (Supplementary
information, Fig. S1). To develop such trimeric ACE2 proteins, we chose a C-terminal domain of T4 fibritin (foldon),8 or a three helix bundle (3HB).9 We also analyzed the reported SARS-CoVs
spike protein structures and estimated that distances between RBDs on the same spike protein could range from 60 to 100 Å when these three RBDs are in the three up conformation.7,10 We then
chose a flexible (GGGGS)5 linker, or a more rigid (EAAAK)5 linker, to construct four trimeric ACE2 proteins: ACE2-flexible-3HB, ACE2-rigid-3HB, ACE2-flexible-foldon and ACE2-rigid-foldon.11
In addition, we constructed two trimeric ACE2 proteins with a short linker GGGS (ACE2-short-3HB, ACE2-short-foldon) and a monomeric ACE2 as control (Supplementary information, Fig. S2).
Binding affinities between ACE2 proteins and the prefusion stabilized trimeric SARS-CoV-2 spike protein ectodomain (S-ECD) were determined by ELISA (Supplementary information, Fig. S3).7
ACE2 monomer binds S-ECD with IC50 of 27 nM. Significant enhancement of the binding affinity was observed for trimeric ACE2 proteins, and the rigid linker constructs showed the highest
binding affinities, including ACE2-rigid-3HB and ACE2-rigid-foldon, which both bound S-ECD with IC50 of 30 pM. Trimeric ACE2 proteins with short linkers had lower binding affinities
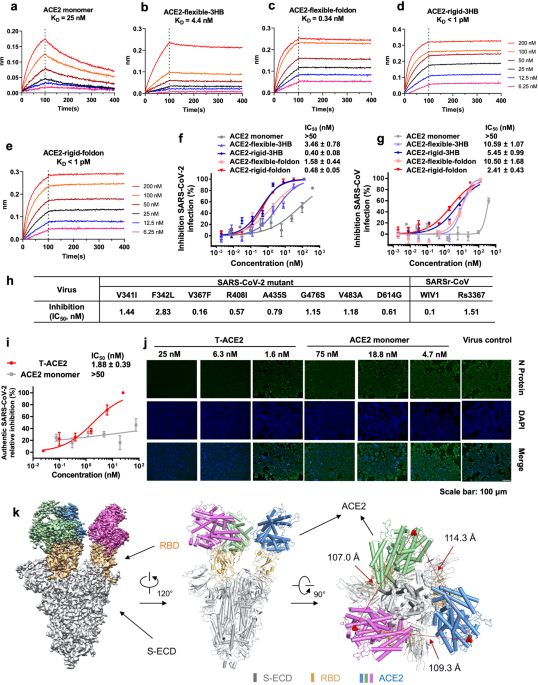
(Supplementary information, Fig. S3). Biolayer interferometry was used to further analyze the binding affinity between ACE2 proteins and S-ECD (Fig. 1a–e). Trimeric ACE2 proteins again
exhibited dramatically increased binding affinities. KD for ACE2-flexible-3HB/S-ECD was 4.4 nM while KD for ACE2-flexible-foldon/S-ECD went down to 0.34 nM. Both ACE2-rigid-3HB and
ACE2-rigid-foldon bound S-ECD extremely tight, with KD < 1 pM. Further decreasing loading of S-ECD on streptavidin sensors did not affect ACE2 proteins binding, suggesting intramolecular
avidity binding between trimeric ACE2s and S-ECD (Supplementary information, Fig. S4). Next, we assessed the inhibitory activities of these trimeric ACE2 decoy proteins using SARS-CoV-2 and
SARS-CoV pseudotyped viruses (Fig. 1f, g). ACE2 monomer can only inhibit SARS-CoV-2 pseudotyped virus at high concentration with IC50 > 50 nM. As expected, trimeric ACE2 with flexible
linkers showed much better inhibitory activities. ACE2-flexible-3HB inhibited SARS-CoV-2 infection with IC50 of 3.46 nM, while ACE2-flexible-foldon had better inhibitory activity with IC50
of 1.58 nM. Rigid-linker trimeric ACE2 proteins again displayed the highest inhibitory activities. ACE2-rigid-3HB and ACE2-rigid-foldon showed similar IC50 of 0.40 and 0.48 nM, respectively.
Short-linker trimeric ACE2 proteins did not show potent inhibitory activities (Supplementary information, Fig. S5). Similar results were observed with SARS-CoV pseudotyped virus inhibition,
and ACE2-rigid-foldon had the best inhibitory activity with IC50 of 2.41 nM. Thus, the ACE2-rigid-foldon construct is the most potent trimeric ACE2, which is designated T-ACE2 hereafter. We
then asked whether T-ACE2 could also inhibit SARS-CoV-2 mutants and SARSr-CoVs. We tested T-ACE2 inhibitory activities on eight naturally occurring SARS-CoV-2 mutants, including seven RBD
domain mutants12 and D614G mutant, and two SARSr-CoVs (WIV1 and Rs3367). We found that T-ACE2 could potently inhibit all these viruses at low concentrations (nM to sub-nM IC50, Fig. 1h;
Supplementary information, Fig. S6). The observed inhibitory activities prompt us to speculate that T-ACE2 might inhibit more SARS-CoV-2 novel mutants. We further tested the inhibition of
authentic SARS-CoV-2 infection by T-ACE2 (Fig. 1i, j). Importantly, we found that T-ACE2 could also potently inhibit SARS-CoV-2 infection with IC50 of 1.88 nM, which is consistent with our
binding affinity and pseudotyped virus inhibition results. We hypothesized that properly designed trimeric ACE2 might engage more than one RBDs from the trimeric spike protein and thus
dramatically increase binding affinity through avidity effect. To further validate this hypothesis, we determined the T-ACE2/S-ECD complex structure using cryo-electron microscopy (cryo-EM)
(Supplementary information, Figs. S7–S9 and Table S1). Strikingly, in the complex, the spike protein adopts only one conformation: the “three-up” RBD conformation. The complex has a nearly
perfect three-fold symmetry. Most importantly, all three RBDs bind to three ACE2s simultaneously (Fig. 1k). The binding interactions between ACE2 and RBD essentially agree with previous
studies, and the three copies from the complex are well aligned (Supplementary information, Figs. S9, S10).13 But our spike protein conformation is very different from the previously
reported prefusion stabilized spike protein structures where only one or no RBD is in the up position. Recent complex structures between SARS-CoV-2 spike protein and ACE2 monomer indicate
that monomer ACE2 binding can induce conformational changes of spike protein and that some spike proteins can have two or three RBDs in the up position to bind up to three ACE2s.14 In our
structure, the unique “three-up” RBD conformation in all the spike proteins should have been induced by our T-ACE2. Whether this T-ACE2-induced spike protein conformation change represents a
transition state during virus infection needs to be further elucidated. Full-length ACE2 protein functions as a dimer,13 the two monomers from this ACE2 dimer are related by two-fold
symmetry, which are situated close in space, with the distance between each D615 being about 53 Å. Thus, the native dimeric ACE2 is unlikely to engage more than one RBD from the same spike
protein without substantial conformational changes. It is however possible that ACE2 dimers on the cell surface might further cluster to induce more RBDs to adopt up conformation and help
virus to transit from the prefusion state to the postfusion state. In summary, we engineered trimeric ACE2 proteins based on wild-type ACE2 and showed that T-ACE2 could bind spike protein
with extremely high affinity to potently inhibit all tested pseudotyped viruses including SARS-CoV-2, SARS-CoV, eight naturally occurring SARS-CoV-2 mutants, two SARSr-CoVs as well as
authentic SARS-CoV-2. Carrying these advancements a few steps beyond, the modular design of T-ACE2 demonstrates that other oligomerization motifs and linkers could be further explored to
improve the properties of T-ACE2 or higher oligomeric ACE2s. We believe that T-ACE2 represents a promising class of proteins to broadly inhibit SARS-CoVs and to treat virus-infected
patients. Finally, the high binding affinity between T-ACE2 and spike protein suggests that T-ACE2 could also be useful for virus detection. The fact that T-ACE2 was engineered based on
native ACE2 sequence renders such detection methods potential application for all SARS-CoVs and related viruses. REFERENCES * Zhou, P. et al. _Nature_ 579, 270–273 (2020). Article CAS
Google Scholar * Korber, B. et al. _Cell_ 182, 812–827 (2020). Article CAS Google Scholar * Imai, Y. et al. _Nature_ 436, 112–116 (2005). Article CAS Google Scholar * Kuba, K. et al.
_Nat. Med._ 11, 875–879 (2005). Article CAS Google Scholar * Hofmann, H. et al. _Biochem. Biophys. Res. Commun._ 319, 1216–1221 (2004). Article CAS Google Scholar * Monteil, V. et al.
_Cell_ 181, 905–913 (2020). Article CAS Google Scholar * Wrapp, D. et al. _Science_ 367, 1260–1263 (2020). Article CAS Google Scholar * Yang, X. et al. _J. Virol._ 76, 4634–4642
(2002). Article CAS Google Scholar * Fletcher, J. M. et al. _ACS Synth. Biol._ 1, 240–250 (2012). Article CAS Google Scholar * Kirchdoerfer, R. N. et al. _Sci. Rep._ 8, 15701 (2018).
Article Google Scholar * Chen, X., Zaro, J. L. & Shen, W. C. _Adv. Drug Deliv. Rev._ 65, 1357–1369 (2013). Article CAS Google Scholar * Li, Q. et al. _Cell_ 182, 1284–1294 (2020).
Article CAS Google Scholar * Yan, R. et al. _Science_ 367, 1444–1448 (2020). Article CAS Google Scholar * Zhou, T. et al. _bioRxiv_ https://doi.org/10.1101/2020.07.04.187989 (2020).
Article PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank the Cryo-EM facility, Supercomputer Center of Westlake University. We thank the Core Facility
of Microbiology and Parasitology (SHMC) and the Biosafety Level 3 Laboratory at Shanghai Medical College of Fudan University, especially Di Qu, Yutang Li, Qian Wang, Zhiping Sun and
Chengjian Gu for continuous support. We thank Tian Li, Xingyue Bao, Mohamad Sawan, Dan Ma and Huaizong Shen for helping protein and cell preparations. We thank Peihui Wang for sharing ACE2
plasmid. This work was supported by Westlake Education Foundation and Tencent Foundation, the National Megaprojects of China for Major Infectious Diseases (2018ZX10301403 to L.L.), the
Natural Science Foundation of China (22077104, 32022037, 31971123, 81920108015, and 31930059), the Key R&D Program of Zhejiang Province (2020C04001), and the SARS-CoV-2 emergency project
of the Science and Technology Department of Zhejiang Province (2020C03129), and the Leading Innovative and Entrepreneur Team Introduction Program of Hangzhou. AUTHOR INFORMATION Author
notes * These authors contributed equally: Liang Guo, Wenwen Bi, Xinling Wang, Wei Xu, Renhong Yan, Yuanyuan Zhang * These authors jointly supervised this work: Youhua Xie, Qiang Zhou, Lu
Lu, Bobo Dang AUTHORS AND AFFILIATIONS * Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China Liang Guo,
Wenwen Bi, Renhong Yan, Yuanyuan Zhang, Kai Zhao, Yaning Li, Mingfeng Zhang, Qiang Zhou & Bobo Dang * Center for Infectious Disease Research, Westlake Laboratory of Life Sciences and
Biomedicine, Hangzhou, Zhejiang, 310024, China Liang Guo, Wenwen Bi, Renhong Yan, Yuanyuan Zhang, Kai Zhao, Yaning Li, Mingfeng Zhang, Qiang Zhou & Bobo Dang * Institute of Biology,
Westlake Institute for Advanced Study, Hangzhou, Zhejiang 310024, China Liang Guo, Wenwen Bi, Renhong Yan, Yuanyuan Zhang, Kai Zhao, Yaning Li, Mingfeng Zhang, Qiang Zhou & Bobo Dang *
Key Laboratory of Medical Molecular Virology (MOE/NHC/CAMS), School of Basic Medical Sciences and Biosafety Level 3 Laboratory, Fudan University, Shanghai, 200032, China Xinling Wang, Wei
Xu, Xia Cai, Shibo Jiang, Youhua Xie & Lu Lu Authors * Liang Guo View author publications You can also search for this author inPubMed Google Scholar * Wenwen Bi View author publications
You can also search for this author inPubMed Google Scholar * Xinling Wang View author publications You can also search for this author inPubMed Google Scholar * Wei Xu View author
publications You can also search for this author inPubMed Google Scholar * Renhong Yan View author publications You can also search for this author inPubMed Google Scholar * Yuanyuan Zhang
View author publications You can also search for this author inPubMed Google Scholar * Kai Zhao View author publications You can also search for this author inPubMed Google Scholar * Yaning
Li View author publications You can also search for this author inPubMed Google Scholar * Mingfeng Zhang View author publications You can also search for this author inPubMed Google Scholar
* Xia Cai View author publications You can also search for this author inPubMed Google Scholar * Shibo Jiang View author publications You can also search for this author inPubMed Google
Scholar * Youhua Xie View author publications You can also search for this author inPubMed Google Scholar * Qiang Zhou View author publications You can also search for this author inPubMed
Google Scholar * Lu Lu View author publications You can also search for this author inPubMed Google Scholar * Bobo Dang View author publications You can also search for this author inPubMed
Google Scholar CONTRIBUTIONS B.D. conceived the project. L.G., K.Z. and M.Z. performed the biochemical experiments, W.B., X.W., W.X, X.C., and Y.L. performed the viral experiments. R.Y.,
Y.Z. and Y.L. solved the cryo-EM structure. B.D., L.L., Q.Z., Y.X. and S.J. designed and supervised the experiments, interpreted the data, wrote and revised the manuscript. CORRESPONDING
AUTHORS Correspondence to Qiang Zhou, Lu Lu or Bobo Dang. ETHICS DECLARATIONS COMPETING INTERESTS B.D., L.G. and W.B. are the inventors on a provisional patent filed by the Westlake
University. The other authors declare no competing interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Guo, L., Bi, W., Wang, X. _et al._ Engineered trimeric ACE2 binds viral spike protein and locks it in “Three-up” conformation to potently inhibit SARS-CoV-2 infection. _Cell Res_
31, 98–100 (2021). https://doi.org/10.1038/s41422-020-00438-w Download citation * Received: 26 September 2020 * Accepted: 25 October 2020 * Published: 11 November 2020 * Issue Date: January
2021 * DOI: https://doi.org/10.1038/s41422-020-00438-w SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
The better sister finale unveils who killed adam with twist in last scenes_WARNING: THIS ARTICLE CONTAINS MAJOR SPOILERS FROM THE BETTER SISTER. _ The gripping series The Better Sister has been ...
Bbc axes 'most perfect film ever' and fans have just days to watchDevotees of the film will be disheartened to discover that it's bidding farewell to the BBC, with only a few days r...
'i'm a gastroenterologist - this is the best time for your first bowel movement'A gastroenterologist has suggested the best time for someone to have their first bowel movement of the day as awareness ...
103-year-old Surrey bowling green could be demolished for state-of-the-art health centreNews103-year-old Surrey bowling green could be demolished for state-of-the-art health centreThe offer has been described...
Surrey town named 'best place in england' for new businessesGuildford has been named England's best place for new businesses according to new data, and has managed to beat out...
Latests News
Engineered trimeric ace2 binds viral spike protein and locks it in “three-up” conformation to potently inhibit sars-cov-2 infectionDear Editor, Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ...
Kaagaz movie review: treading on fragile groundSTORY: In order to keep his band-baja business afloat, Bharat Lal (Pankaj Tripathi) approaches a local bank in Uttar Pra...
Brittany mahomes posts new video from las vegas before super bowl 2024The Super Bowl 2024 stage is set and Brittany Mahomes is ready for kickoff in Las Vegas. Taking to her Instagram Stories...
Weekend storm alerts: french weather outlook august 24 - 25PREDICTIONS FOR TEMPERATURE, SUN, STORM, RAIN AND MORE Storms are set to cover much of France this weekend, as a brief b...
Il-2 engineered mscs rescue t cells in tumoursIL-2 is a powerful growth factor for T cells. New work shows that immune checkpoint blockade depends upon the presence o...