Lipid production in nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator
Lipid production in nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
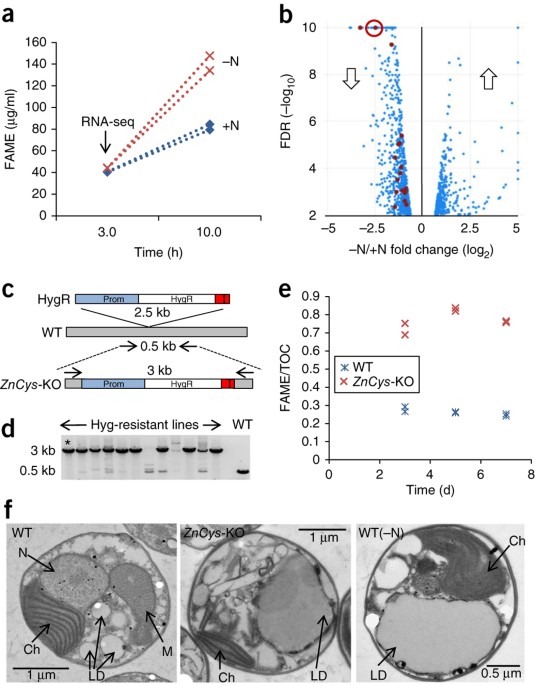
ABSTRACT Lipid production in the industrial microalga _Nannochloropsis gaditana_ exceeds that of model algal species and can be maximized by nutrient starvation in batch culture. However,
starvation halts growth, thereby decreasing productivity. Efforts to engineer _N. gaditana_ strains that can accumulate biomass and overproduce lipids have previously met with little
success. We identified 20 transcription factors as putative negative regulators of lipid production by using RNA-seq analysis of _N. gaditana_ during nitrogen deprivation. Application of a
CRISPR–Cas9 reverse-genetics pipeline enabled insertional mutagenesis of 18 of these 20 transcription factors. Knocking out a homolog of fungal Zn(II)2Cys6-encoding genes improved
partitioning of total carbon to lipids from 20% (wild type) to 40–55% (mutant) in nutrient-replete conditions. Knockout mutants grew poorly, but attenuation of Zn(II)2Cys6 expression yielded
strains producing twice as much lipid (∼5.0 g m−2 d−1) as that in the wild type (∼2.5 g m−2 d−1) under semicontinuous growth conditions and had little effect on growth. Access through your
institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio
journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per
year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
CHARACTERIZATION AND RNA-SEQ TRANSCRIPTOMIC ANALYSIS OF A _SCENEDESMUS OBLIQNUS_ MUTANT WITH ENHANCED PHOTOSYNTHESIS EFFICIENCY AND LIPID PRODUCTIVITY Article Open access 03 June 2021 IRON
RESCUES GLUCOSE-MEDIATED PHOTOSYNTHESIS REPRESSION DURING LIPID ACCUMULATION IN THE GREEN ALGA _CHROMOCHLORIS ZOFINGIENSIS_ Article Open access 18 July 2024 A GUANIDINE-DEGRADING ENZYME
CONTROLS GENOMIC STABILITY OF ETHYLENE-PRODUCING CYANOBACTERIA Article Open access 26 August 2021 ACCESSION CODES PRIMARY ACCESSIONS SEQUENCE READ ARCHIVE * SRP096211 REFERENCES * Goncalves,
E.C., Wilkie, A.C., Kirst, M. & Rathinasabapathi, B. Metabolic regulation of triacylglycerol accumulation in the green algae: identification of potential targets for engineering to
improve oil yield. _Plant Biotechnol. J._ 14, 1649–1660 (2016). Article CAS Google Scholar * Hu, Q. et al. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives
and advances. _Plant J._ 54, 621–639 (2008). Article CAS Google Scholar * Vasudevan, V. et al. Environmental performance of algal biofuel technology options. _Environ. Sci. Technol._ 46,
2451–2459 (2012). Article CAS Google Scholar * Wijffels, R.H. & Barbosa, M.J. An outlook on microalgal biofuels. _Science_ 329, 796–799 (2010). Article CAS Google Scholar * Ngan,
C.Y. et al. Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. _Nature Plants_ 1, 15107 (2015). Article CAS Google Scholar * Goold, H.D. et al.
Whole genome re-sequencing identifies a quantitative trait locus repressing carbon reserve accumulation during optimal growth in _Chlamydomonas reinhardtii_. _Sci. Rep._ 6, 25209 (2016).
Article CAS Google Scholar * Schulz-Raffelt, M. et al. Hyper-accumulation of starch and oil in a _Chlamydomonas_ mutant affected in a plant-specific DYRK kinase. _Biotechnol. Biofuels_ 9,
55 (2016). Article Google Scholar * Trentacoste, E.M. et al. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. _Proc. Natl.
Acad. Sci. USA_ 110, 19748–19753 (2013). Article CAS Google Scholar * Daboussi, F. et al. Genome engineering empowers the diatom _Phaeodactylum tricornutum_ for biotechnology. _Nat.
Commun._ 5, 3831 (2014). Article CAS Google Scholar * Levitan, O., Dinamarca, J., Zelzion, E., Gorbunov, M.Y. & Falkowski, P.G. An RNA interference knock-down of nitrate reductase
enhances lipid biosynthesis in the diatom _Phaeodactylum tricornutum_. _Plant J._ 84, 963–973 (2015). Article CAS Google Scholar * Radakovits, R. et al. Draft genome sequence and genetic
transformation of the oleaginous alga _Nannochloropis gaditana_. _Nat. Commun._ 3, 686 (2012). Article Google Scholar * Griffiths, M.J. & Harrison, S.T.L. Lipid productivity as a key
characteristic for choosing algal species for biodiesel production. _J. Appl. Phycol._ 21, 493–507 (2009). Article CAS Google Scholar * Ma, X.N., Chen, T.P., Yang, B., Liu, J. & Chen,
F. Lipid production from _Nannochloropsis_. _Mar. Drugs_ 14, 61 (2016). Article Google Scholar * Kilian, O., Benemann, C.S., Niyogi, K.K. & Vick, B. High-efficiency homologous
recombination in the oil-producing alga _Nannochloropsis_ sp. _Proc. Natl. Acad. Sci. USA_ 108, 21265–21269 (2011). Article CAS Google Scholar * Rodolfi, L. et al. Microalgae for oil:
strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. _Biotechnol. Bioeng._ 102, 100–112 (2009). Article CAS Google Scholar * Sukenik,
A. et al. Photosynthetic perfromance of outdoor _Nannochloropsis_ mass cultures under a wide range of environmental conditions. _Aquat. Microb. Ecol._ 56, 297–308 (2009). Article Google
Scholar * Blazeck, J. et al. Harnessing _Yarrowia lipolytica_ lipogenesis to create a platform for lipid and biofuel production. _Nat. Commun._ 5, 3131 (2014). Article Google Scholar *
Friedlander, J. et al. Engineering of a high lipid producing _Yarrowia lipolytica_ strain. _Biotechnol. Biofuels_ 9, 77 (2016). Article Google Scholar * Zienkiewicz, K., Du, Z.Y., Ma, W.,
Vollheyde, K. & Benning, C. Stress-induced neutral lipid biosynthesis in microalgae: molecular, cellular and physiological insights. _Biochim. Biophys. Acta_ 1861 9 Pt B, 1269–1281
(2016). Article CAS Google Scholar * Boussiba, S.V.A., Cohen, Z., Avissar, Y. & Richmond, A. Lipid and biomass production by the halotolerant microalga _Nannochloropsis salina_.
_Biomass_ 12, 37–47 (1987). Article CAS Google Scholar * Li, J. et al. Choreography of transcriptomes and lipidomes of nannochloropsis reveals the mechanisms of oil synthesis in
microalgae. _Plant Cell_ 26, 1645–1665 (2014). Article CAS Google Scholar * Corteggiani Carpinelli, E. et al. Chromosome scale genome assembly and transcriptome profiling of
_Nannochloropsis gaditana_ in nitrogen depletion. _Mol. Plant_ 7, 323–335 (2014). Article CAS Google Scholar * Pérez-Rodríguez, P. et al. PlnTFDB: updated content and new features of the
plant transcription factor database. _Nucleic Acids Res._ 38, D822–D827 (2010). Article Google Scholar * Cerutti, H., Ma, X., Msanne, J. & Repas, T. RNA-mediated silencing in algae:
biological roles and tools for analysis of gene function. _Eukaryot. Cell_ 10, 1164–1172 (2011). Article CAS Google Scholar * Wang, Q. et al. Genome editing of model oleaginous microalgae
_Nannochloropsis_ spp. by CRISPR/Cas9. _Plant J._ 88, 1071–1081 (2016). Article CAS Google Scholar * Kennerdell, J.R. & Carthew, R.W. Heritable gene silencing in _Drosophila_ using
double-stranded RNA. _Nat. Biotechnol._ 18, 896–898 (2000). Article CAS Google Scholar * Vieler, A., Brubaker, S.B., Vick, B. & Benning, C. A lipid droplet protein of
_Nannochloropsis_ with functions partially analogous to plant oleosins. _Plant Physiol._ 158, 1562–1569 (2012). Article CAS Google Scholar * Xiao, Y., Zhang, J., Cui, J., Feng, Y. &
Cui, Q. Metabolic profiles of _Nannochloropsis oceanica_ IMET1 under nitrogen-deficiency stress. _Bioresour. Technol._ 130, 731–738 (2013). Article CAS Google Scholar * Schmollinger, S.
et al. Nitrogen-sparing mechanisms in _Chlamydomonas_ affect the transcriptome, the proteome, and photosynthetic metabolism. _Plant Cell_ 26, 1410–1435 (2014). Article CAS Google Scholar
* Gargouri, M. et al. Identification of regulatory network hubs that control lipid metabolism in _Chlamydomonas reinhardtii_. _J. Exp. Bot._ 66, 4551–4566 (2015). Article CAS Google
Scholar * Hu, J. et al. Genome-wide identification of transcription factors and transcription-factor binding sites in oleaginous microalgae _Nannochloropsis_. _Sci. Rep._ 4, 5454 (2014).
Article Google Scholar * Xu, P., Qiao, K., Ahn, W.S. & Stephanopoulos, G. Engineering _Yarrowia lipolytica_ as a platform for synthesis of drop-in transportation fuels and
oleochemicals. _Proc. Natl. Acad. Sci. USA_ 113, 10848–10853 (2016). Article CAS Google Scholar * Biddy, M.J. et al. The techno-economic basis for coproduct manufacturing to enable
hydrocarbon fuel production from lignocellulosic biomass. _ACS Sustain. Chem.& Eng._ 4, 3196–3211 (2016). Article CAS Google Scholar * Davis, R., Aden, A. & Pienkos, P.T.
Techno-economic analysis of autotrophic microalgae for fuel production. _Appl. Energy_ 88, 3524–3531 (2011). Article Google Scholar * Mortazavi, A., Williams, B.A., McCue, K., Schaeffer,
L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. _Nat. Methods_ 5, 621–628 (2008). Article CAS Google Scholar * Li, B. & Dewey, C.N. RSEM: accurate
transcript quantification from RNA-Seq data with or without a reference genome. _BMC Bioinformatics_ 12, 323 (2011). Article CAS Google Scholar * McCarthy, D.J., Chen, Y. & Smyth,
G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. _Nucleic Acids Res._ 40, 4288–4297 (2012). Article CAS Google Scholar *
Young, M.D., Wakefield, M.J., Smyth, G.K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. _Genome Biol._ 11, R14 (2010). Article Google Scholar * Shen,
B. et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. _Cell Res._ 23, 720–723 (2013). Article CAS Google Scholar * Cho, S.W., Kim, S., Kim, J.M. & Kim,
J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. _Nat. Biotechnol._ 31, 230–232 (2013). Article CAS Google Scholar * Mali, P. et al. RNA-guided human
genome engineering via Cas9. _Science_ 339, 823–826 (2013). Article CAS Google Scholar * Bligh, E.G. & Dyer, W.J. A rapid method of total lipid extraction and purification. _Can. J.
Biochem. Physiol._ 37, 911–917 (1959). Article CAS Google Scholar * Ruiz-Matute, A.I., Hernández-Hernández, O., Rodríguez-Sánchez, S., Sanz, M.L. & Martínez-Castro, I. Derivatization
of carbohydrates for GC and GC-MS analyses. _J. Chromatogr. B Analyt. Technol. Biomed. Life Sci._ 879, 1226–1240 (2011). Article CAS Google Scholar * Kaspar, H. _Amino acid analysis in
biological fluids by GC-MS_. Dr. rer. nat. thesis, Univ. Regensburg (2009). Google Scholar * Lichtenthaler, H.K. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes.
_Methods Enzymol._ 148, 32 (1987). Google Scholar * Littler, M.M. & Arnold, K.E. In _Handbook of Phycological Methods. Ecological Field Methods: Macroalgae_ (eds. Littler M.M. &
Littler, D.S.) 349–375 (Cambridge University Press, 1985). Download references ACKNOWLEDGEMENTS This work was funded by ExxonMobil and Synthetic Genomics, Inc. We thank A. Withrow (Center
for Advanced Microscopy at Michigan State University) for producing the TEM images and C. Packard, B. Scherer, E. Wang and the rest of the analytical team at SGI for processing FAME and TOC
samples. This work is dedicated to our colleague Tom Carlson, who passed away during the preparation of this manuscript. AUTHOR INFORMATION Author notes * Tom J Carlson: Deceased. AUTHORS
AND AFFILIATIONS * Synthetic Genomics Inc., La Jolla, California, USA Imad Ajjawi, John Verruto, Moena Aqui, Leah B Soriaga, Jennifer Coppersmith, Kathleen Kwok, Luke Peach, Elizabeth
Orchard, Ryan Kalb, Weidong Xu, Tom J Carlson, Kristie Francis, Katie Konigsfeld, Judit Bartalis, Andrew Schultz, William Lambert, Ariel S Schwartz, Robert Brown & Eric R Moellering
Authors * Imad Ajjawi View author publications You can also search for this author inPubMed Google Scholar * John Verruto View author publications You can also search for this author
inPubMed Google Scholar * Moena Aqui View author publications You can also search for this author inPubMed Google Scholar * Leah B Soriaga View author publications You can also search for
this author inPubMed Google Scholar * Jennifer Coppersmith View author publications You can also search for this author inPubMed Google Scholar * Kathleen Kwok View author publications You
can also search for this author inPubMed Google Scholar * Luke Peach View author publications You can also search for this author inPubMed Google Scholar * Elizabeth Orchard View author
publications You can also search for this author inPubMed Google Scholar * Ryan Kalb View author publications You can also search for this author inPubMed Google Scholar * Weidong Xu View
author publications You can also search for this author inPubMed Google Scholar * Tom J Carlson View author publications You can also search for this author inPubMed Google Scholar * Kristie
Francis View author publications You can also search for this author inPubMed Google Scholar * Katie Konigsfeld View author publications You can also search for this author inPubMed Google
Scholar * Judit Bartalis View author publications You can also search for this author inPubMed Google Scholar * Andrew Schultz View author publications You can also search for this author
inPubMed Google Scholar * William Lambert View author publications You can also search for this author inPubMed Google Scholar * Ariel S Schwartz View author publications You can also search
for this author inPubMed Google Scholar * Robert Brown View author publications You can also search for this author inPubMed Google Scholar * Eric R Moellering View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS I.A. and E.R.M. conceived the study and designed experiments. R.B. provided technical advice. E.O. and R.K. designed the
productivity assays. L.B.S. and A.S.S. performed computational and bioinformatics analyses. M.A., J.V., J.C., L.P., J.B., A.S., W.X., T.J.C., K.F., W.L., K. Kwok and K. Konigsfeld performed
the experiments. I.A. and E.R.M. wrote the manuscript with support from all authors. CORRESPONDING AUTHORS Correspondence to Imad Ajjawi or Eric R Moellering. ETHICS DECLARATIONS COMPETING
INTERESTS I.A., J.V., M.A., J.C., K. Kwok, L.P., E.O., R.K., K. Konigsfeld, A.S., W.L., R.B. and E.R.M. are employees of Synthetic Genomics, Inc. Synthetic genomics has filed patents related
to this work, with I.A., J.V., M.A., L.B.S. and E.R.M. listed as inventors. INTEGRATED SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 TAG INDUCTION OF _ZNCYS_-KO FOR CULTURES GROWN IN
BATCH MODE ON SM-NO3−. A) FAME/TOC comparison of knockout lines for 18 –N down-regulated transcription regulators (see Supplementary Table 1 for full gene annotation information). FAME/TOC
values represent the average and standard deviation of 3 time-points from biological duplicates during batch growth. The knockout line in gene Naga_100104g1g (referred to as _ZnCys_-KO)
selected for further study is shown in red. B) Confirmation of the increased FAME/TOC ratio in a second Cas9-mediated _ZnCys_-KO line 2; the _ZnCys_-KO line 1 was used in the remainder of
the study and is referred to as _ZnCys_-KO. C) FAME (mg/L) and D) TOC values (mg/L) of _ZnCys_-KO and WT grown in batch mode on SM-NO3- (n=2) E) FAME profiles (as mol % of total FAME)
showing most abundant fatty acid species for _ZnCys_-KO in N-replete conditions and WT in +N and -N. C# indicates the fatty acid carbon chain length and:# indicates the number of double
bonds. F) TAG content normalized to TOC (g/g) as determined by LC-MS. See Materials and Methods for further details on growth conditions. SUPPLEMENTARY FIGURE 2 INITIAL BATCH-MODE ASSESSMENT
OF _ZNCYS_-ATTENUATED LINES (_ZNCYS_-BASH-3, _ZNCYS_-BASH-12 AND _ZNCYS_-RNAI-7) GROWN IN NITRATE-REPLETE MEDIUM (SM-NO3−). A) FAME (mg/L) and B) TOC (mg/L) measurements corresponding to
days 3, 5 and 7 of the screen. TOC productivity values displayed in Fig 2C were derived from these values. SUPPLEMENTARY FIGURE 3 PRODUCTIVITY ASSESSMENT OF _ZNCYS_-KO AND _ZNCYS_-RNAI-7
GROWN IN SEMICONTINUOUS MODE ON NITRATE-RICH MEDIUM (SM-NO3−). Daily (A) FAME (mg/L), (B) TOC (mg/L), and (C) C/N values derived from cellular N-content. (D) FAME and E) TOC productivities
(g/m2/day) for WT and _ZnCys_-RNAi-7 calculated for the entire 13-day assay. _ZnCys_-KO failed to reach steady-state at a 30 % daily dilution scheme and essentially washed away as the run
progressed, therefore lipid and biomass productivity values were not calculated for this line (N/A, not available). Due to the severe growth defect of _ZnCys_-KO on medium containing nitrate
as the sole N source, this strain was scaled up in SM-NH4+/NO3- to obtain enough biomass for the assay, but grown on SM-NO3- medium for the duration of semi-continuous productivity
assessment. SUPPLEMENTARY FIGURE 4 PRODUCTIVITY ASSESSMENT OF _ZNCYS_ MUTANTS GROWN IN SEMICONTINUOUS MODE FOR 8 D ON NO3−-RICH MEDIUM (SM-NO3−). A) Daily FAME and B) TOC (mg/L) measurements
for _ZnCys_ mutants (_ZnCys_-RNAi-7, _ZnCys_-BASH-12 and _ZnCys_-BASH-3) compared to their parental lines Ng-CAS9+ and WT. Productivity values displayed in Fig 3A were derived from these
values. Error bars represent standard deviations for 3 biological replicates (n=3). See Materials and Methods section for a detailed description on the assay and productivity calculations.
C) Cell counts for various strains. Shown is the average and standard deviation of biological triplicate cultures for three consecutive days under semi-continuous growth (N = 9;
corresponding to days 6 through 8 in Figure S4A). SUPPLEMENTARY FIGURE 5 REPRESSION OF THE LIPID-ACCUMULATION PHENOTYPE OF _ZNCYS_-KO BY NH4+ SUPPLEMENTATION. Daily A) FAME (mg/L), B) TOC
(mg/L) and C) FAME/TOC values of _ZnCys_-KO and WT grown in batch mode on medium supplemented with NH4+ (SM-NH4+/NO3-). Error bars are standard deviations of 2 biological replicates.
SUPPLEMENTARY FIGURE 6 DOSE-DEPENDENT EFFECT OF CYCLOHEXIMIDE TREATMENT ON FAME/TOC. Cultures were grown as in Fig. 4, and treated with the indicated amount of cycloheximide at 0 h. FAME/TOC
measurements were taken 48 h after treatment. SUPPLEMENTARY FIGURE 7 DIEL LIGHT PROFILES USED IN THIS STUDY. Incident irradiance profiles for batch growth assessment (A), and the
Semi-Continuous Productivity Assay (B). SUPPLEMENTARY FIGURE 8 DIAGRAMS OF VECTOR CONSTRUCTS USED IN THIS STUDY. Vector used to generate the _Nannochloropsis_ Cas9 expression strain Ng-Cas9+
(A), and the hygromycin resistance cassette used for generating Cas9-mediated insertional mutants in the Ng-Cas9+ background (B). The coding sequences (yellow) for BSD (blasticidin
deaminase), Cas9, and GFP, are driven by the TCT_P, RPL24_P, and 4AIII_P endogenous promoters, and terminated by the EIF3_T, FRD_T, and GNPDA_T endogenous terminator sequences, respectively.
See Supplementary Table 7 for a further description of gene sources for promoter and terminator elements. SUPPLEMENTARY FIGURE 9 SELECTION OF THE NG-CAS9+ EDITOR LINE AND VALIDATION OF
TRANSGENIC CAS9 PROTEIN EXPRESSION. (A) Flow cytometry histogram (using the Accuri c6 cytometer) showing fluorescence in the FL1-A channel to detect GFP fluorescence. Wild-type fluorescence
is shown in the black trace, whereas Ng-CAS9+ is shown in red. Histograms are drawn in the Accuri c6 Sampler software using data collected from 50,000 events. (B) Western blot detection of
transgenic Cas9 protein in the Ng-CAS9+ editor line. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–9, Supplementary Tables 1–9 and Supplementary Notes 1–3
(PDF 1403 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Ajjawi, I., Verruto, J., Aqui, M. _et al._ Lipid production in _Nannochloropsis gaditana_
is doubled by decreasing expression of a single transcriptional regulator. _Nat Biotechnol_ 35, 647–652 (2017). https://doi.org/10.1038/nbt.3865 Download citation * Received: 08 November
2016 * Accepted: 05 April 2017 * Published: 19 June 2017 * Issue Date: July 2017 * DOI: https://doi.org/10.1038/nbt.3865 SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative
Trending News
Two cases of kappa covid-19 variant detected in uttar pradeshAfter two cases of the Delta plus strain were found in Uttar Pradesh's Deoria and Gorakhpur, two cases of the Kappa...
Turning point uk was destined to fail. Here's why | thearticleThe arrival of Turning Point UK (TPUK), a British offshoot of the American right-wing youth pressure group has, all thin...
Express. Home of the Daily and Sunday Express.Live-event Fury as Labour set to take credit for slashing migrant numbers - despite it being Tory plan Follow our live b...
Zombies: why they matter in the philosophy of mind | thearticleWho (perhaps I should say “ what”) is your favourite zombie? Which dextrously eccentric eater of flesh (there are no veg...
Caregiver tips: conquer hard to reach zippersDo you have certain tricks up your caregiver sleeve that make everyday tasks a bit easier? Share your creative, low-cost...
Latests News
Lipid production in nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulatorABSTRACT Lipid production in the industrial microalga _Nannochloropsis gaditana_ exceeds that of model algal species and...
404 errorFête des Voisins: 10 million neighbours in France to meet this Friday 2025 marks the 25th edition of solidarity festival...
Nine accused of politkovskaya killingProsecutors have charged nine people, including a senior Federal Security Service officer, over their alleged involvemen...
Strictly come dancing tour backlash: performers slammed for ‘mocking'A video shared by current host of the tour, Ore Oduba, has sparked outrage once again. The video shows Neil and Katya J...
There are no standards in public life any more | thearticleFor most of his working life, Boris Johnson has behaved as though rules and social norms were meant for others, not him....