Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens
Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The immune system influences the fate of developing cancers by not only functioning as a tumour promoter that facilitates cellular transformation, promotes tumour growth and sculpts
tumour cell immunogenicity1,2,3,4,5,6, but also as an extrinsic tumour suppressor that either destroys developing tumours or restrains their expansion1,2,7. Yet, clinically apparent cancers
still arise in immunocompetent individuals in part as a consequence of cancer-induced immunosuppression. In many individuals, immunosuppression is mediated by cytotoxic T-lymphocyte
associated antigen-4 (CTLA-4) and programmed death-1 (PD-1), two immunomodulatory receptors expressed on T cells8,9. Monoclonal-antibody-based therapies targeting CTLA-4 and/or PD-1
(checkpoint blockade) have yielded significant clinical benefits—including durable responses—to patients with different malignancies10,11,12,13. However, little is known about the identity
of the tumour antigens that function as the targets of T cells activated by checkpoint blockade immunotherapy and whether these antigens can be used to generate vaccines that are highly
tumour-specific. Here we use genomics and bioinformatics approaches to identify tumour-specific mutant proteins as a major class of T-cell rejection antigens following anti-PD-1 and/or
anti-CTLA-4 therapy of mice bearing progressively growing sarcomas, and we show that therapeutic synthetic long-peptide vaccines incorporating these mutant epitopes induce tumour rejection
comparably to checkpoint blockade immunotherapy. Although mutant tumour-antigen-specific T cells are present in progressively growing tumours, they are reactivated following treatment with
anti-PD-1 and/or anti-CTLA-4 and display some overlapping but mostly treatment-specific transcriptional profiles, rendering them capable of mediating tumour rejection. These results reveal
that tumour-specific mutant antigens are not only important targets of checkpoint blockade therapy, but they can also be used to develop personalized cancer-specific vaccines and to probe
the mechanistic underpinnings of different checkpoint blockade treatments. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this
article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in
* Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS IL-18BP IS A SECRETED IMMUNE CHECKPOINT AND BARRIER TO IL-18
IMMUNOTHERAPY Article 24 June 2020 ANTIGEN PRESENTATION IN CANCER: INSIGHTS INTO TUMOUR IMMUNOGENICITY AND IMMUNE EVASION Article 09 March 2021 REGULATORY T CELL-MEDIATED IMMUNOSUPPRESSION
ORCHESTRATED BY CANCER: TOWARDS AN IMMUNO-GENOMIC PARADIGM FOR PRECISION MEDICINE Article 29 February 2024 ACCESSION CODES PRIMARY ACCESSIONS GENE EXPRESSION OMNIBUS * GSE62771 DATA DEPOSITS
RNA-sequencing data are available at Gene Expression Omnibus (GEO) repository at http://www.ncbi.nlm.nih.gov/geo/ (accession number GSE62771). REFERENCES * Shankaran, V. et al. IFNγ and
lymphocytes prevent primary tumour development and shape tumour immunogenicity. _Nature_ 410, 1107–1111 (2001) ADS CAS PubMed Google Scholar * Dunn, G. P., Bruce, A. T., Ikeda, H., Old,
L. J. & Schreiber, R. D. Cancer immunoediting: from immunosurveillance to tumor escape. _Nature Immunol._ 3, 991–998 (2002) CAS Google Scholar * Mantovani, A., Allavena, P., Sica, A.
& Balkwill, F. Cancer-related inflammation. _Nature_ 454, 436–444 (2008) ADS CAS PubMed Google Scholar * Grivennikov, S. I., Greten, F. R. & Karin, M. Immunity, inflammation, and
cancer. _Cell_ 140, 883–899 (2010) CAS PubMed PubMed Central Google Scholar * Trinchieri, G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. _Annu. Rev.
Immunol._ 30, 677–706 (2012) CAS PubMed Google Scholar * Coussens, L. M., Zitvogel, L. & Palucka, A. K. Neutralizing tumor-promoting chronic inflammation: a magic bullet? _Science_
339, 286–291 (2013) ADS CAS PubMed PubMed Central Google Scholar * Koebel, C. M. et al. Adaptive immunity maintains occult cancer in an equilibrium state. _Nature_ 450, 903–907 (2007)
ADS CAS PubMed Google Scholar * Quezada, S. A., Peggs, K. S., Simpson, T. R. & Allison, J. P. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication.
_Immunol. Rev._ 241, 104–118 (2011) CAS PubMed PubMed Central Google Scholar * Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. _Nature Rev. Cancer_ 12, 252–264
(2012) CAS Google Scholar * Wolchok, J. D. et al. Nivolumab plus ipilimumab in advanced melanoma. _N. Engl. J. Med._ 369, 122–133 (2013) CAS PubMed PubMed Central Google Scholar *
Hamid, O. et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. _N. Engl. J. Med._ 369, 134–144 (2013) CAS PubMed PubMed Central Google Scholar * Topalian, S. L.
et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. _N. Engl. J. Med._ 366, 2443–2454 (2012) CAS PubMed PubMed Central Google Scholar * Hodi, F. S. et al.
Improved survival with ipilimumab in patients with metastatic melanoma. _N. Engl. J. Med._ 363, 711–723 (2010) CAS PubMed PubMed Central Google Scholar * Matsushita, H. et al. Cancer
exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. _Nature_ 482, 400–404 (2012) ADS CAS PubMed PubMed Central Google Scholar * Paul, S. et al. HLA class I
alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. _J. Immunol._ 191, 5831–5839 (2013) CAS PubMed Google Scholar * West, E. E. et al.
Tight regulation of memory CD8+ T cells limits their effectiveness during sustained high viral load. _Immunity_ 35, 285–298 (2011) CAS PubMed PubMed Central Google Scholar * Wherry, E.
J. T cell exhaustion. _Nature Immunol._ 12, 492–499 (2011) CAS Google Scholar * Castle, J. C. et al. Exploiting the mutanome for tumor vaccination. _Cancer Res._ 72, 1081–1091 (2012) CAS
PubMed Google Scholar * Robbins, P. F. et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. _Nature Med._ 19,
747–752 (2013) CAS PubMed Google Scholar * Fritsch, E. F. et al. HLA-binding properties of tumor neoepitopes in humans. _Cancer Immunol. Res._ 2, 522–529 (2014) CAS PubMed PubMed
Central Google Scholar * van Rooij, N. et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. _J. Clin. Oncol._ 31, e439–e442
(2013) PubMed Google Scholar * Curran, M. A., Montalvo, W., Yagita, H. & Allison, J. P. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and
myeloid cells within B16 melanoma tumors. _Proc. Natl Acad. Sci. USA_ 107, 4275–4280 (2010) ADS CAS PubMed PubMed Central Google Scholar * Brunner, M. C. et al. CTLA-4-mediated
inhibition of early events of T cell proliferation. _J. Immunol._ 162, 5813–5820 (1999) CAS PubMed Google Scholar * Keir, M. E., Butte, M. J., Freeman, G. J. & Sharpe, A. H. PD-1 and
its ligands in tolerance and immunity. _Annu. Rev. Immunol._ 26, 677–704 (2008) CAS PubMed Google Scholar * Okazaki, T., Chikuma, S., Iwai, Y., Fagarasan, S. & Honjo, T. A rheostat
for immune responses: the unique properties of PD-1 and their advantages for clinical application. _Nature Immunol._ 14, 1212–1218 (2013) CAS Google Scholar * Duraiswamy, J., Kaluza, K.
M., Freeman, G. J. & Coukos, G. Dual blockade of PD-1 and CTLA-4 combined with tumor vaccine effectively restores T-cell rejection function in tumors. _Cancer Res._ 73, 3591–3603 (2013)
CAS PubMed PubMed Central Google Scholar * Spiotto, M. T., Rowley, D. A. & Schreiber, H. Bystander elimination of antigen loss variants in established tumors. _Nature Med._ 10,
294–298 (2004) CAS PubMed Google Scholar * Wolkers, M. C., Brouwenstijn, N., Bakker, A. H., Toebes, M. & Schumacher, T. N. Antigen bias in T cell cross-priming. _Science_ 304,
1314–1317 (2004) ADS CAS PubMed Google Scholar * Corbière, V. et al. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. _Cancer Res._ 71,
1253–1262 (2011) PubMed Google Scholar * Tran, E. et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. _Science_ 344, 641–645 (2014) ADS
CAS PubMed PubMed Central Google Scholar * Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. _Science_ 322, 1097–1100
(2008) ADS CAS PubMed PubMed Central Google Scholar * Peters, B. & Sette, A. Generating quantitative models describing the sequence specificity of biological processes with the
stabilized matrix method. _BMC Bioinformatics_ 6, 132 (2005) PubMed PubMed Central Google Scholar * Lundegaard, C. et al. NetMHC-3.0: accurate web accessible predictions of human, mouse
and monkey MHC class I affinities for peptides of length 8-11. _Nucleic Acids Res._ 36, W509–W512 (2008) CAS PubMed PubMed Central Google Scholar * Hoof, I. et al. NetMHCpan, a method
for MHC class I binding prediction beyond humans. _Immunogenetics_ 61, 1–13 (2009) CAS PubMed Google Scholar * Nielsen, M., Lundegaard, C., Lund, O. & Kesmir, C. The role of the
proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. _Immunogenetics_ 57, 33–41 (2005) CAS PubMed Google Scholar *
Esquivel, F., Yewdell, J. & Bennink, J. RMA/S cells present endogenously synthesized cytosolic proteins to class I-restricted cytotoxic T lymphocytes. _J. Exp. Med._ 175, 163–168 (1992)
CAS PubMed Google Scholar * Toebes, M. et al. Design and use of conditional MHC class I ligands. _Nature Med._ 12, 246–251 (2006) CAS PubMed Google Scholar * Andersen, R. S. et al.
Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. _Nature Protocols_ 7, 891–902 (2012) CAS PubMed Google Scholar * Kvistborg, P. et al.
TIL therapy broadens the tumor-reactive CD8+ T cell compartment in melanoma patients. _OncoImmunology_ 1, 409–418 (2012) PubMed PubMed Central Google Scholar * Kowalewski, D. J. &
Stevanovic, S. Biochemical large-scale identification of MHC class I ligands. _Methods Mol. Biol._ 960, 145–157 (2013) CAS PubMed Google Scholar * Thommen, D. S. et al. Two preferentially
expressed proteins protect vascular endothelial cells from an attack by peptide-specific CTL. _J. Immunol._ 188, 5283–5292 (2012) CAS PubMed Google Scholar * Domon, B. & Aebersold,
R. Options and considerations when selecting a quantitative proteomics strategy. _Nature Biotechnol._ 28, 710–721 (2010) CAS Google Scholar * MacLean, B. et al. Skyline: an open source
document editor for creating and analyzing targeted proteomics experiments. _Bioinformatics_ 26, 966–968 (2010) CAS PubMed PubMed Central Google Scholar * Lange, V., Picotti, P., Domon,
B. & Aebersold, R. Selected reaction monitoring for quantitative proteomics: a tutorial. _Mol. Syst. Biol._ 4, 222 (2008) PubMed PubMed Central Google Scholar * Escher, C. et al.
Using iRT, a normalized retention time for more targeted measurement of peptides. _Proteomics_ 12, 1111–1121 (2012) CAS PubMed PubMed Central Google Scholar * Subramanian, A. et al. Gene
set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. _Proc. Natl Acad. Sci. USA_ 102, 15545–15550 (2005) ADS CAS PubMed PubMed Central
Google Scholar Download references ACKNOWLEDGEMENTS We are grateful to K. Murphy for the _Batf3__−/−_ mice, T. Hansen for providing MHC class I antibodies and the H-2Kb construct, D.
Fremont for the human β2m construct, and the National Institutes of Health (NIH) Tetramer Core Facility for producing MHC class I tetramers. We also thank R. Ahmed and M. Hashimoto for the
multiplex staining strategy used to define functional and dysfunctional T cells. We thank A. Bensimon, O. Schubert and P. Kouvonen for instrument maintenance and for technical support with
the mass spectrometry measurements and R. Vanganipuram, M. Selby and J. Valle for generating and supplying anti-PD-1 and anti-CTLA-4 in endotoxin-free sterile form. We also thank K. Sheehan,
P. Allen, G. Dunn and R. Chan for constructive criticisms and comments, all members of the Schreiber laboratory for discussions, and the many members of The Genome Institute at Washington
University School of Medicine. We would also like to thank W. Song for his assistance with the bioinformatics approaches, P. Kvistborg for assistance with tetramer combinatorial coding, and
Christopher Nelson for advice with peptide-MHC monomer purification. This work was supported by grants to R.D.S. from the National Cancer Institute (RO1 CA043059, U01 CA141541), the Cancer
Research Institute and the WWW.W Foundation; to R.D.S. and W.E.G. from The Siteman Cancer Center/Barnes-Jewish Hospital (Cancer Frontier Fund); to W.E.G. from Susan G. Komen for the Cure
(Promise grant); to E.R.M. from the National Human Genome Research Institute; to G.J.F. from the National Institute of Health (P50 CA101942, P01 AI054456, P50 CA101942); to A.H.S. from the
National Institute of Health (P50 CA101942); and to T.N.S. from the Dutch Cancer Society (Queen Wilhelmina Research Award). E.C. is supported by a Marie Curie Intra-European Fellowship
within the Seventh Framework Programme of the European Community for Research. M.M.G. was supported by a postdoctoral training grant (T32 CA00954729) from the National Cancer Institute and
is currently supported by a postdoctoral training grant (Irvington Postdoctoral Fellowship) from the Cancer Research Institute. Aspects of studies at Washington University were performed
with assistance by the Immunomonitoring Laboratory of the Center for Human Immunology and Immunotherapy Programs and the Siteman Comprehensive Cancer Center. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Department of Pathology and Immunology, Washington University School of Medicine, 660 South Euclid Avenue, St Louis, Missouri 63110, USA, Matthew M. Gubin, Jeffrey P. Ward,
Takuro Noguchi, Yulia Ivanova, Cora D. Arthur, Matthew D. Vesely, Samuel S. K. Lam, Erika L. Pearce, Maxim N. Artyomov & Robert D. Schreiber * Department of Surgery, Washington
University School of Medicine, 660 South Euclid Avenue, St Louis, Missouri 63110, USA, Xiuli Zhang & William E. Gillanders * Department of Immunology, Institute of Cell Biology, and
German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ) Partner Site Tübingen, Auf der Morgenstelle 15, 72076 Tübingen, Germany, Heiko Schuster & Hans-Georg Rammensee *
Department of Biology, Institute of Molecular Systems Biology, ETH Zurich, 8093 Zurich, Switzerland, Etienne Caron & Ruedi Aebersold * Department of Medicine, Division of Oncology,
Washington University School of Medicine, 660 South Euclid Avenue, St Louis, Missouri 63110, USA, Jeffrey P. Ward * The Genome Institute, Washington University School of Medicine, 4444
Forest Park Avenue, St Louis, Missouri 63108, USA, Jasreet Hundal & Elaine R. Mardis * ISA Therapeutics B.V., 2333 CH Leiden, The Netherlands, Willem-Jan Krebber, Gwenn E. Mulder &
Cornelis J. M. Melief * Division of Immunology, The Netherlands Cancer Institute, 1066 CX Amsterdam, The Netherlands, Mireille Toebes & Ton N. Schumacher * Bristol-Myers Squibb, 700 Bay
Road, Redwood City, 94063, California, USA Alan J. Korman * Department of Immunology, The University of Texas MD Anderson Cancer Center, Houston, 77030, Texas, USA James P. Allison *
Department of Medical Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, 02115, Massachusetts, USA Gordon J. Freeman * Department of Microbiology and Immunobiology,
Harvard Medical School, Boston, 02115, Massachusetts, USA Arlene H. Sharpe * Faculty of Science, University of Zurich, Zurich, 8093 Zurich, Switzerland, Ruedi Aebersold * Department of
Immunohematology and Blood Transfusion, Leiden University Medical Center, 2333ZA Leiden, The Netherlands, Cornelis J. M. Melief * Department of Genetics, Washington University School of
Medicine, 660 South Euclid Avenue, St Louis, Missouri 63110, USA, Elaine R. Mardis Authors * Matthew M. Gubin View author publications You can also search for this author inPubMed Google
Scholar * Xiuli Zhang View author publications You can also search for this author inPubMed Google Scholar * Heiko Schuster View author publications You can also search for this author
inPubMed Google Scholar * Etienne Caron View author publications You can also search for this author inPubMed Google Scholar * Jeffrey P. Ward View author publications You can also search
for this author inPubMed Google Scholar * Takuro Noguchi View author publications You can also search for this author inPubMed Google Scholar * Yulia Ivanova View author publications You can
also search for this author inPubMed Google Scholar * Jasreet Hundal View author publications You can also search for this author inPubMed Google Scholar * Cora D. Arthur View author
publications You can also search for this author inPubMed Google Scholar * Willem-Jan Krebber View author publications You can also search for this author inPubMed Google Scholar * Gwenn E.
Mulder View author publications You can also search for this author inPubMed Google Scholar * Mireille Toebes View author publications You can also search for this author inPubMed Google
Scholar * Matthew D. Vesely View author publications You can also search for this author inPubMed Google Scholar * Samuel S. K. Lam View author publications You can also search for this
author inPubMed Google Scholar * Alan J. Korman View author publications You can also search for this author inPubMed Google Scholar * James P. Allison View author publications You can also
search for this author inPubMed Google Scholar * Gordon J. Freeman View author publications You can also search for this author inPubMed Google Scholar * Arlene H. Sharpe View author
publications You can also search for this author inPubMed Google Scholar * Erika L. Pearce View author publications You can also search for this author inPubMed Google Scholar * Ton N.
Schumacher View author publications You can also search for this author inPubMed Google Scholar * Ruedi Aebersold View author publications You can also search for this author inPubMed Google
Scholar * Hans-Georg Rammensee View author publications You can also search for this author inPubMed Google Scholar * Cornelis J. M. Melief View author publications You can also search for
this author inPubMed Google Scholar * Elaine R. Mardis View author publications You can also search for this author inPubMed Google Scholar * William E. Gillanders View author publications
You can also search for this author inPubMed Google Scholar * Maxim N. Artyomov View author publications You can also search for this author inPubMed Google Scholar * Robert D. Schreiber
View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.M.G. and R.D.S. were involved in all aspects of this study including planning and
performing experiments, analysing and interpreting data, and writing the manuscript. X.Z. performed peptide binding experiments, helped design and perform the vaccine experiments. H.S.,
E.C., R.A. and H.-G.R. planned and performed the mass spectrometry analyses, interpreted the data and were involved in writing the manuscript. T.N., J.P.W., C.D.A., M.D.V., S.S.K.L. and
E.L.P., participated in assessing the phenotypes of the tumour-specific T-cell lines, interpreting the data and in writing the manuscript. M.T. helped generate MHC class I multimers. A.J.K.,
J.P.A., G.J.F. and A.H.S. provided mAbs, helped plan the checkpoint blockade therapy experiments, and contributed to writing the manuscript. T.N.S. helped generate MHC class I multimers,
analysed data and was involved in writing the manuscript. W.-J.K., G.E.M. and C.J.M.M. produced and purified the synthetic long peptides, participated in the planning of the vaccine
experiments, analysed data and were involved in writing the manuscript. J.H. and E.R.M. were responsible for genomic analyses and epitope prediction and participated in writing the
manuscript. W.E.G. contributed to the design and analysis of peptide binding and vaccine experiments and in writing the manuscript. Y.I. and M.N.A were responsible for optimizing the epitope
prediction method, performing the RNA-sequencing analyses, analysing data and writing the manuscript. R.D.S. oversaw all the work performed. CORRESPONDING AUTHOR Correspondence to Robert D.
Schreiber. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIGURE 1 INNATE AND ADAPTIVE IMMUNE
COMPONENTS ARE REQUIRED FOR REJECTION OF D42M1-T3 AFTER CHECKPOINT BLOCKADE THERAPY. A, Cohorts of _Rag2_−/−, _Batf3_−/− or wild-type mice were treated with control mAb, anti-CD4, anti-CD8α
or anti-IFN-γ mAbs and then were injected with 1 × 106 d42m1-T3 tumour cells subcutaneously and subsequently treated with anti-CTLA-4 on days 3, 6 and 9 post-transplant. B, C, d42m1-T3 (B)
or F244 (C) tumour cells were injected subcutaneously into wild-type mice (_n_ = 5) that were subsequently treated with anti-PD-1 on days 3, 6, and 9. Fifty days after tumours were rejected,
mice were rechallenged with d42m1-T3 or F244 tumour cells. Data are presented as average tumour diameter ± s.e.m. of 5 mice per group and are representative of at least two independent
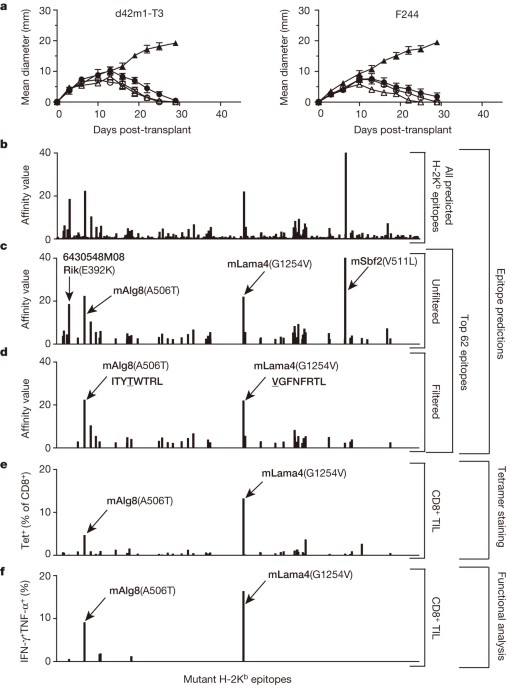
experiments. EXTENDED DATA FIGURE 2 H-2DB MUTANT EPITOPES OF D42M1-T3 TUMOURS. A, Missense mutations in d42m1-T3 were subjected to _in silico_ analysis for the potential to form
H-2Db-binding epitopes using three epitope prediction algorithms. The median predicted epitope-binding affinity for each peptide was calculated and expressed as ‘median affinity value’ where
affinity value = 1/IC50 × 100. Predicted epitopes are arrayed along the _x_-axis in alphabetical order based on their protein of origin. B, Unfiltered median affinity values for the four
predicted H-2Db epitopes. C, Median affinity values of remaining two H-2Db epitopes after filtering. D, Tetramer staining of CD8+ TIL from tumour-bearing mice treated with anti-PD-1 using
H-2Db tetramers loaded with top 4 H-2Db synthetic peptides. E, IFN-γ and TNF-α intracellular cytokine staining of CD8+ TIL from tumour-bearing mice treated with anti-PD-1 immunotherapy
following co-culture with naive irradiated splenocytes pulsed with the top four H-2Db synthetic peptides added at 1 μM final concentration. Data are presented as per cent of CD8+ TIL
positive for IFN-γ, TNF-α or both cytokines. Data are representative of two independent experiments. EXTENDED DATA FIGURE 3 MLAMA4 AND MALG8 STIMULATE CD8+ T CELL LINES GENERATED AGAINST
D42M1-T3 FOLLOWING ANTI-PD-1 IMMUNOTHERAPY. A, CD8+ T cell lines generated from splenocytes of individual d42m1-T3-tumour-bearing mice that rejected their tumours after anti-PD-1 therapy
were incubated with irradiated d42m1-T3 tumour cells (or F244 tumour cells) treated with blocking mAb specific for H-2Kb, and/or H-2Db and IFN-γ production was quantitated. Data are
presented as means ± s.e.m. and are representative of two independent experiments. Samples were compared using an unpaired, two-tailed Student’s _t_ test (***_P_ < 0.001). B, IFN-γ
release by the CTL 74 T cell line following co-culture with naive irradiated splenocytes pulsed with the top 62 H-2Kb synthetic peptides added at 1 μM final concentration. EXTENDED DATA
FIGURE 4 MLAMA4 AND MALG8 BIND H-2KB AND STIMULATE CD8+ T CELL LINES GENERATED AGAINST D42M1-T3 FOLLOWING ANTI-PD-1 IMMUNOTHERAPY. A, IFN-γ release by CTL 62, CTL 73 or CTL 74 T cell lines
following stimulation with naive irradiated splenocytes pulsed with wild-type or mutant forms of Lama4 or Alg8 peptides. B, RMA-S cells were incubated with 8 amino acid peptides of mLama4 or
mAlg8 and surface expression of H-2Kb or H-2Db was assessed by flow cytometry. Mean fluorescent intensity of H-2Kb and H-2Db was expressed as peptide binding score. Data presented are
representative of at least two independent experiments. EXTENDED DATA FIGURE 5 IDENTIFICATION OF A PEPTIDE BOUND TO H-2KB ON D42M1-T3 TUMOUR CELLS CORRESPONDING TO MUTANT LAMA4. A,
Identification of the peptide, VGFNFRTL, corresponding to mLama4 by discovery mass spectrometry. B, Validation of the mLama4 peptide using an isotope-labelled synthetic peptide (VGFNFRTL
(13C6, 15N1)). EXTENDED DATA FIGURE 6 GENERATION OF SRM ASSAY LIBRARY FOR THE DETECTION OF MUTANT H-2KB PEPTIDES ON D42M1-T3. A, SRM transitions were optimized for 51 of the 62 top predicted
H-2Kb peptides. The 51 peptides chosen were selected based on having physiochemical properties that would allow their detection by mass spectrometry if present. Only Lama4 and Alg8 are
shown here for simplicity. The 51 peptides were synthesized and LC-tandem mass spectrometry acquisition was performed on each peptide to determine the best collision energy and to obtain the
full fragment ion spectrum (left panel); three to seven of the highest intensity peaks were selected to be built into SRM transitions. Optimal SRM transitions displayed as extracted ion
chromatograms are shown (right panel). Q1–Q3 transitions are indicated in parenthesis. The mutated amino acid in the peptide sequence is marked in red. B, F244 tumour cells, which lack the
mLama4 and mAlg8 d42m1-T3 epitope, lack detectable mLama4 or mAlg8 in complex with H-2Kb as assessed by SRM. EXTENDED DATA FIGURE 7 CD8+ T CELLS SPECIFIC FOR MUTANT FORMS OF LAMA4 AND ALG8
INFILTRATE D42M1-T3, BUT NOT F244, TUMOURS. A, Detection of tumour-infiltrating mLama4- or mAlg8-specific T cells infiltrating d42m1-T3 or F244 tumours of mice treated with anti-PD-1.
Tumours were harvested on day 12 post-transplant. Cells were gated on live CD45+ and CD8α+ tumour-infiltrating lymphocytes. Detection of mLama4- or mAlg8-specific T cells was achieved by
staining with peptide-MHC H-2Kb PE-labelled tetramers. Data are representative of at least five independent experiments. B, Detection of mLama4-specific tumour-infiltrating T cells from
tumour-bearing mice treated with anti-PD-1, anti-CTLA-4, both anti-PD-1 plus anti-CTLA-4 or control mAb. Detection of mLama4-specific T cells was achieved by staining with mLama4-MHC H-2Kb
PE-labelled tetramers. Data presented are plotted as the mean mLama4 tetramer-positive as a percent of CD8α+ tumour-infiltrating cells and are representative of at least three independent
experiments. EXTENDED DATA FIGURE 8 MALG8 AND MLAMA4 SLP VACCINE CONTROL D42M1-T3 TUMOUR OUTGROWTH WHEN ADMINISTERED THERAPEUTICALLY OR PROPHYLACTICALLY. A, Tumour growth of d42m1-T3 tumours
from mice therapeutically vaccinated with mLama4 and mAlg8 SLP plus poly(I:C), HPV control SLP plus poly(I:C) or poly(I:C) alone. Data shown are mean ± s.e.m. Mutant Lama4 and mAlg8 SLP
vaccine group was compared to HPV control SLP vaccine group using an unpaired, two-tailed Student’s _t_ test (*_P_ < 0.05 and **_P_ < 0.01). B, Kaplan–Meier survival curves of
d42m1-T3-tumour-bearing mice (7 per group) prophylactically vaccinated with SLP vaccines plus poly(I:C). mLama4 plus mAlg8 compared to HPV control: _P_ = 0.0003 (log-rank (Mantel–Cox) test).
Representative of two independent experiments. C, Cumulative number of mice (7–10 per group) from at least two independent experiments rejecting d42m1-T3 or F244 tumours as a consequence of
SLP or minimal epitope peptide prophylactic vaccination. EXTENDED DATA FIGURE 9 DETECTION OF TIM-3, LAG-3, IFN-Γ AND TNF-Α EXPRESSION IN TUMOUR-INFILTRATING CD8+ T CELLS. A, Representative
histogram of TIM-3 or LAG-3 expression on mLama4-specific CD8+ tumour-infiltrating T cells from tumour-bearing mice treated with anti-PD-1, anti-CTLA-4, both anti-PD-1 and anti-CTLA-4 or
control mAbs. B, TIM-3 and LAG-3 are reduced in mAlg8-specific CD8+ TIL from tumour-bearing mice treated with anti-PD-1, anti-CTLA-4, or both anti-PD-1 and anti-CTLA-4 compared to mice
treated with control mAb. _n_ = 5 mice per group pooled. Data are presented as mean ± s.e.m. of at least three independent experiments. Samples were compared using an unpaired, two-tailed
Student’s _t_ test (*_P_ < 0.05, **_P_ < 0.01). C, Representative dot plots of IFN-γ and TNF-α stained CD8+ tumour-infiltrating T cells from tumour-bearing mice following treatment
with anti-PD-1, anti-CTLA-4, both anti-PD-1 and anti-CTLA-4 or control mAbs. Data presented are representative of at least three independent experiments. SUPPLEMENTARY INFORMATION
SUPPLEMENTARY DATA 1 This file contains MS traces for each of the mutant H-2Kb d42m1-T3 epitopes tested by targeted MS. (PDF 1891 kb) SUPPLEMENTARY INFORMATION This file contains
Supplementary Tables 1-3. Supplementary Table 1 contains a complete list of d42m1-T3 H-2Kb-bound peptides identified by discovery MS. Supplementary Table 2 shows all differentially expressed
genes in mLama4-tetramer-positive CD8+ TILs from mice treated with checkpoint blockade therapy compared to mLama4-tetramer-positive CD8+ TILs from control mice. Supplementary Table 3 shows
differentially regulated pathways (GSEA pathway analysis) in mLama4-tetramer-positive CD8+ TILs from mice treated with checkpoint blockade therapy compared to mLama4-tetramer-positive CD8+
TILs from control mice. (PDF 3029 kb) POWERPOINT SLIDES POWERPOINT SLIDE FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 POWERPOINT SLIDE FOR FIG. 4 RIGHTS AND PERMISSIONS
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gubin, M., Zhang, X., Schuster, H. _et al._ Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens.
_Nature_ 515, 577–581 (2014). https://doi.org/10.1038/nature13988 Download citation * Received: 14 May 2014 * Accepted: 22 October 2014 * Published: 26 November 2014 * Issue Date: 27
November 2014 * DOI: https://doi.org/10.1038/nature13988 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
1190 effect of human growth hormone (hgh) treatment on fat metabolismABSTRACT Administration of pharmacological quantities of hGH to humans and animals causes a brisk rise in serum free fat...
Refining pain management in mice by comparing multimodal analgesia and nsaid monotherapy for neurosurgical proceduresABSTRACT While neurosurgical interventions are frequently used in laboratory mice, refinement efforts to optimize analge...
A divalent sirna chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous systemABSTRACT Sustained silencing of gene expression throughout the brain using small interfering RNAs (siRNAs) has not been ...
Antitumor efficacy of aav-mediated systemic delivery of interferon-βABSTRACT Type I interferons (_α_/_β_) have significant antitumor activity although their short half-life and systemic si...
Reading che guevara in his own wordsChe Guevara was killed on 9 October 1967, but new books by him are still appearing. He was a prolific writer and the Cen...
Latests News
Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigensABSTRACT The immune system influences the fate of developing cancers by not only functioning as a tumour promoter that f...
Unlock some paywalls with a news aggregator subscriptionMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...
Poppies a hit with young generationWhat was becoming an event confined to the older generation with memories of the Second World War is now engaging the wh...
Journal club | NatureAccess through your institution Buy or subscribe A BIOCHEMIST IS EXCITED BY A UNIVERSAL GLUE FOR MOLECULAR BIOLOGY. Inve...
Deadline’s bafta awards live blogThe EE BAFTA Film Awards are underway here at Southbank Centre’s Royal Festival Hall in London. The ceremony is on a del...