The screening of the 3′UTR sequence of LRRK2 identified an association between the rs66737902 polymorphism and Parkinson’s disease
The screening of the 3′UTR sequence of LRRK2 identified an association between the rs66737902 polymorphism and Parkinson’s disease"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Mutations in the leucine-rich repeat kinase 2 gene (LRRK2) are the most common genetic determinants of familial and sporadic Parkinson’s disease (PD). Most of the mutational screenings
analyzed the exon-coding sequence. Our aim was to determine whether LRRK2 3′ untranslated region (UTR) variants were associated with the risk of developing PD in a large cohort of patients
(n=743) and controls (n=523) from Spain. We identified a total of 12 3′UTR variants (two new). Single-nucleotide polymorphism (SNP) rs66737902 C allele was overrepresented in patients
(P=0.005; odds ratio=1.47). This SNP was in linkage disequilibrium with the p.R1441G mutation, but the association remained significant among mutation-negative cases. We found a significant
lower level of the LRRK2 transcript in the Substantia nigra (SN) of PD postmortem donors (n=9) who were rs66737902 C carriers (P=0.01). This SNP was predicted to affect a binding site for
miR-138-2-3p. We showed that this microRNA was expressed in all the SN samples. In conclusion, we found a significant association between SNP rs66737902 and the risk of developing PD. This
effect on PD risk could be explained by differences in LRRK2 transcript levels between the two alleles.
Parkinson’s disease (PD) affects 1–2% of people older than 60 years.1 In addition to rare mutations that have been linked to familial forms of PD, DNA polymorphisms at several genes could
contribute to the risk of developing sporadic PD. The leucine-rich repeat kinase 2 gene (LRRK2, PARK8) has been linked to both sporadic and familial PD.2, 3, 4, 5 The LRRK2 p.G2019S mutation
accounts for 30–40% of PD cases among North African Berbers patients and Ashkenazi Jews and has also been found in 5% of Spanish patients.6 The p.R1441G mutation is also common in Spain,
with a frequency in PD cases close to 8% in some regions.7 The LRRK2 gene encodes a protein with multiple functional domains, including leucine-rich repeats, a GTPase domain and a kinase
domain, which, in addition to the brain, is expressed in multiple tissues.8 The mutations found in PD patients disrupt the enzymatic activities of the protein by affecting the kinase
(p.G2019S) or the GTPase (p.R1441G/C) activities.9, 10 Some studies also reported differences in LRRK2 expression between brains from PD cases compared with those from controls.11, 12, 13
Most of the mutational screenings were focused on the LRRK2 coding exons, whereas little is known about the contribution of 3′UTR variants to the risk for PD. However, several studies have
provided strong evidence of the role of this gene region on the regulation of LRRK2 expression through microRNA binding, and the control of cellular pathways in which this gene is
implicated. Among others, pathogenic LRRK2 antagonizes let-7 and miR-184-3p in Drosophila, leading to the overproduction of proteins implicated in cell cycle regulation and survival.14 It
was also reported that miR-205 is able to suppress the expression of LRRK2 by targeting a conserved binding site in the 3′UTR region.15 Our first aim was to characterize the contribution of
the LRRK2 3′UTR variation in the development of PD.
The study involved a total of 743 PD patients and 523 healthy controls, all Iberians from the region of Asturias (Northern Spain). The main characteristics of these cohorts are summarized in
Table 1 and were previously reported.16 The LRRK2 3′UTR was amplified in six overlapping fragments (Supplementary Table 1), which were subjected to single strand conformation analysis and
direct sequencing to identify the nucleotide changes. We found a total of 12 variants (Supplementary Table 2), and only the SNP rs66737902 was linked to the risk of developing PD: allele
C-carriers were frequent among the patients (P=0.005; odds ratio=1.47, 95% confidence interval (CI)=1.12–1.92) (Table 2). The sample size was enough for a statistical power >80%. The
difference remained significant after correcting for age and gender (P=0.01; odds ratio=1.47, 95% CI=1.09–1.97). To our knowledge, the association between PD and rs66737902 has not been
reported by others who also examined the LRRK2 3′UTR.4, 17 This SNP showed a heterogeneous worldwide frequency distribution, but was absent among the Chinese and Japanese (www.ensembl.org).
Differences in ethnic background could thus explain these discrepancies.
We found a significant linkage disequilibrium between rs66737902 and p.R1441G (rs33939927): a total of 15 patients were heterozygous for this mutation, and 14 and 1 were rs66737902 TC and
TT, respectively. This mutation was not found among the 105 controls who were rs66737902 C carriers. After excluding the 15 p.R1441G mutation carriers, carriers of rs66737902 C remained
significantly increased among the patients. This suggested that the risk effect of this 3′UTR allele could not be fully explained by its linkage disequilibrium with the LRRK2 mutation. In
contrast, most of the p.G2019S mutation carriers (13/16) were homozygous for the rs66737902 TT protective genotype.
We measured the LRRK2 cDNA levels in three brain regions (SN, cerebellum, and occipital cortex) of 9 PD donors kindly provided by the London Neurodegenerative Diseases Brain Bank
(Supplementary Table 3). Tissue processing and expression assays were performed as reported by Cardo et al.16 Briefly, the level of LRRK2 and GAPDH (constitutively expressed normalization
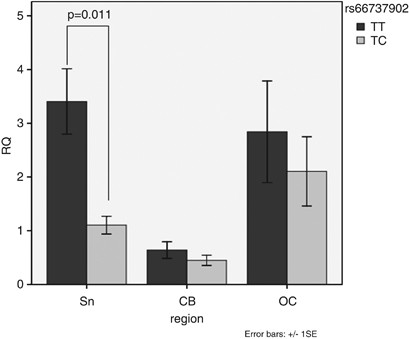
control) cDNA was determined using SYBR Green. The amount of LRRK2 was calculated as the fold difference, 2−ΔΔCt. We found a significant lower mean level of LRRK2 cDNA (P=0.01) in the SN of
rs66737902 TC (n=4) vs TT (n=5) donors (Figure 1). This effect was not seen in the cerebellum and cortex samples. Some studies have found significant reductions of LRRK2 mRNA in the brains
from PD cases compared with controls.11 In the brains of patients with PD, LRRK2 immunoreactivity was reduced in SN neurons, a fact that reflects the disease-associated loss of dopaminergic
neurons in this brain region.12 Another study did not find significant alteration in LRRK2 expression between brain tissue from p.G2019S mutation carriers and those from controls.13 None of
the nine brain donors analyzed was a p.R1441G carrier, and thus we could not examine whether this mutation was also linked to differences in LRRK2 expression. The differences in mRNA
expression between the genotypes could be attributed to its linkage disequilibrium with p.R1441G, but whether they also exist at the protein level can be confirmed only through additional
studies.
Mean LRRK2 expression (cDNA) in the SN, cerebellum (CB) and occipital cortex (OC) of the nine PD donors according to the rs66737902 genotype (4 TC, 5 TT). The level of LRRK2 mRNA was
determined through real-time PCR using ABI7500 equipment (Life Technologies, Carlsbad, CA, USA) and the GAPDH gene was used as the endogenous control for quantification. Each cDNA was
amplified in triplicate using iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) under the following PCR conditions: 95°C for 30 s followed by 40 cycles at 95°C for 15 s and at
58°C for 34 s, and a final melting curve stage. The amount of LRRK2 was calculated as the fold difference 2−ΔΔCt, in which ΔCt=Ct LRRK2−Ct GAPDH; ΔΔCt=ΔCt sample−ΔCt controls mean. LRRK2
primers: ATA TAA AGG CTC GCG CTT CT and TGG CAT TCA CAA AGT GGT AAT (amplified fragment=162 bp). GADPH primers: GGA AGG ACT CAT GAC CAC AG and TTG GCA GGT TTT TCT AGA CGT (amplified
fragment=249 bp).
Finally, an online analysis with two miRNA-site prediction software programs (http://www.microrna.org; http://diana.cslab.ece.ntua.gr/) identified miR-138.2 as a candidate that could bind to
the rs66737902 T sequence (Supplementary Figure 1). We found that this miRNA was expressed in all the nine SN samples (Supplementary Figure 2). Thus, the PD-protective T allele could result
in enhanced miRNA binding and downregulation of the LLRK2 mRNA. However, this would be in opposition to the higher transcript levels among the TT SN samples. Moreover, this binding site was
recognized by only one of the programs and with a low score (Supplementary Figure 1). Although we considered unlikely a direct effect of the rs66737902 SNP through different binding of
miR-138.2 to the two alleles, in vitro studies with neural cells transfected with reporter vectors coupled to 3′UTRs with the two alleles are required in case other studies confirm the
genetic association.
In conclusion, we report the association between the LRRK2 rs66737902 SNP and PD. This effect could be mediated by a significant reduction of gene expression in the SN of individuals with
the risk allele, and require validation in other populations with larger samples of brain tissues.
We thank Dr Claire Troakes and the London Neurodegenerative Diseases Brain Bank for supplying all post mortem brain samples. We also thank the ‘Fundacion Parkinson Asturias’ and ‘Obra Social
Cajastur’ for their support. This work was supported by grants from the Spanish ‘Fondo de Investigaciones Sanitarias-Fondos FEDER’ European Union (FIS 11/0093). LFC is a predoctoral
fellowship from FICYT-Principado de Asturias.
All the authors contributed to this work by recruiting the patients and obtaining the clinical and anthropometric data or by performing the laboratory work. All authors have read and
approved the submission of this manuscript.
Genética Molecular-Laboratorio de Medicina, Hospital Universitario Central de Asturias, Oviedo, Spain
Neurología, Hospitales Universitario Central Asturias and Álvarez Buylla-Mieres, Asturias, Spain
Geriatric Research, Education and Clinical Center, Veterans Affairs Puget Sound Health Care System, Seattle, WA, USA
Department of Neurology, University of Washington School of Medicine, Seattle, WA, USA
Supplementary Information accompanies the paper on Journal of Human Genetics website
Anyone you share the following link with will be able to read this content:
Trending News
Arsenal hero ian wright urges board to sack unai emery after leicesterArsenal hero Ian Wright has urged the Gunners’ board to make a decision following the defeat to Leicester. Arsenal were ...
Eagle Creek Packable Daypack Sale 2021 | The StrategistPhoto-Illustration: retailer With travel restrictions easing up as temperatures warm up, you might be itching to get awa...
Soviet Union - The MIT Press ReaderThe saga of Clarence Hiskey, a chemist employed by the Manhattan Project, and Arthur Adams, a spy-runner, has largely fa...
Pune airport’s new terminal to open tomorrow: here's all you need to know about itPune Airport’s New Terminal To Open Tomorrow: Here's All You Need To Know About It | X/@mohol_murlidhar Costing ₹42...
Tufts university student dies in accidentLocal News MADIE NICPON WAS A MEMBER OF THE CLASS OF 2023. A Tufts University student is dead following an accident on S...
Latests News
The screening of the 3′UTR sequence of LRRK2 identified an association between the rs66737902 polymorphism and Parkinson’s diseaseMutations in the leucine-rich repeat kinase 2 gene (LRRK2) are the most common genetic determinants of familial and spor...
Who experts split on monkeypox emergency ahead of decision – sourcesWHO experts split on monkeypox emergency ahead of decision – sources | WTVB | 1590 AM · 95.5 FM | The Voice of Branch Co...
18 misconceptions about men women need to change* Home * Viral * 18 Misconceptions about men women need to throw out of the window RIGHT NOW! THERE'S A WHOLE LOT O...
In vitro cytocidal effect of l-glutaminase on leukaemic lymphocytesABSTRACT MOUSE lymphomas, such as 6C3HED, undergo complete regression after one or more injections of L-asparaginase1, a...
David Douglas, 1798–1834 | NatureABSTRACT A BOTANICAL collector and explorer in many British territories, David Douglas, the Scottish naturalist, was bor...