Hsa-mir-520d induces hepatoma cells to form normal liver tissues via a stemness-mediated process
Hsa-mir-520d induces hepatoma cells to form normal liver tissues via a stemness-mediated process"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The human ncRNA gene _RGM249_ regulates the extent of differentiation of cancer cells and the conversion of 293FT cells to hiPSCs. To identify the factors underlying this process,
we investigated the effects of lentivirally inducing miR-520d expression in 293FT and HLF cells _in vitro_. Subsequently, we evaluated tumor formation in a xenograft model. Transformed HLF
cells were Oct4 and Nanog positive within 24 h, showed p53 upregulation and hTERT downregulation and mostly lost their migration abilities. After lentiviral infection, the cells were
intraperitoneally injected into mice, resulting in benign teratomas (6%), the absence of tumors (87%) or differentiation into benign liver tissues (7%) at the injection site after 1 month.
We are the first to demonstrate the loss of malignant properties in cancer cells _in vivo_ through the expression of a single microRNA (miRNA). This miRNA successfully converted 293FT and
hepatoma cells to hiPSC-like cells. The regulation of malignancy by miR-520d appears to be through the conversion of cancer cells to normal stem cells, maintaining p53 upregulation. SIMILAR
CONTENT BEING VIEWED BY OTHERS REGENERATION LINKED MIRNA MODIFY TUMOR PHENOTYPE AND CAN ENFORCE MULTI-LINEAGE GROWTH ARREST IN VIVO Article Open access 18 May 2021 SPINK1-INDUCED TUMOR
PLASTICITY PROVIDES A THERAPEUTIC WINDOW FOR CHEMOTHERAPY IN HEPATOCELLULAR CARCINOMA Article Open access 29 November 2023 CRISPR/CAS9-MEDIATED KNOCKOUT OF PIM3 SUPPRESSES TUMORIGENESIS AND
CANCER CELL STEMNESS IN HUMAN HEPATOBLASTOMA CELLS Article 16 April 2021 INTRODUCTION Mature microRNAs (miRNAs; single-stranded 20–23-nucleotide RNA molecules) control the expression of
genes involved in many cellular processes1. miRNAs typically reduce the stability of mRNAs, including those encoding genes that mediate tumorigenic processes, such as apoptosis, cell cycle
regulation, differentiation, inflammation, invasion and stress responses2. Mammalian miRNAs mediate cellular differentiation and reprogramming and play crucial roles in the initiation and
progression of human cancers3. Alterations in miRNA expression can influence tumor growth by targeting and modulating the functional expression of genes that regulate tumor cell apoptosis or
proliferation4. miRNAs can serve as tumor suppressors (suppressor miRs) and/or oncogenes (oncomiRs) and their expression has been found to be dysregulated in many malignancies5. miRNA
targeting is primarily achieved through specific base-pair interactions between the 5′ ends (‘seed’ region) of miRNAs and target sites within the coding and/or untranslated regions (UTRs) of
mRNAs; target sites in the 3'UTR lead to more effective mRNA destabilization6. Because miRNAs frequently target hundreds of mRNAs, miRNA regulatory pathways are complex7. It is
extremely difficult to achieve control of a cancer by manipulating a single factor, because cancer cells easily escape from induced chemical, physical and molecular stresses through
alternative pathways8. However, miRNAs involved in stemness and the benign state through the simultaneous control of multiple pathways could be expected to curatively convert cancer cells9.
Given that the presence or absence of miRNAs plays a critical role in tumorigenic processes and that miRNA expression occurs in a disease-specific manner, miRNAs possess great potential as
therapeutic targets and novel biomarkers10. miRNAs synergistically induce stemness and pluripotency in cancer cells and specifically in 293FT cells11. For example, recent studies in
reprogrammed human pluripotent stem cells have suggested that the elevated expression of miR-302 family members influenced the cell cycle transition toward homogeneous proliferation. _In
vivo_ studies have shown that miR-302 inhibits the tumorigenicity of human pluripotent stem cells (hPSCs) by enhancing multiple G1 phase arrest pathways, rather than by silencing p21Cip112.
Human miR-520d is a minor miRNA that is involved in HER2/neu receptor-related and osteoblast differentiation, although its function in these processes remains unclear13. miR-520d-5p
upregulation was observed to induce suppressive effects and inhibit metastasis when the expression of human _RGM249_ (which is present on 10p15) was abrogated by gene silencing14. Thus,
_RGM249_ was identified as a candidate miRNA precursor gene that might orchestrate the target genes involved in modulating differentiation, proliferation, malignant alteration or stemness.
_RGM249_ is strongly expressed in poorly differentiated or undifferentiated malignant tumor cell lines (e.g., hepatoma, sarcoma, glioblastoma, thyroid cancer and malignant melanoma) and
might play a role in carcinogenesis or the maintenance of differentiation levels. Here we report a novel and striking role for miR-520d-5p in cancer development and stemness in
undifferentiated hepatoma cell lines (HLF). In this study, we also analyzed the metabolomics profiles of miR-520d-5p transfectants to evaluate the reprogramming levels, as metabolite levels
have been reported to play a role in regulating the epigenetic changes that occur during reprogramming15. Furthermore, we examined a key gene that can interact with miR-520d-5p. RESULTS _IN
VITRO_ STUDY OF MIR-520D-5P-LENTIVIRUS-INFECTED HLF HLF cells that were infected with a miR-520d-5p-expressing lentiviral vector (520d-HLF; hsa-miR-520d-5p-overexpressing HLF) were converted
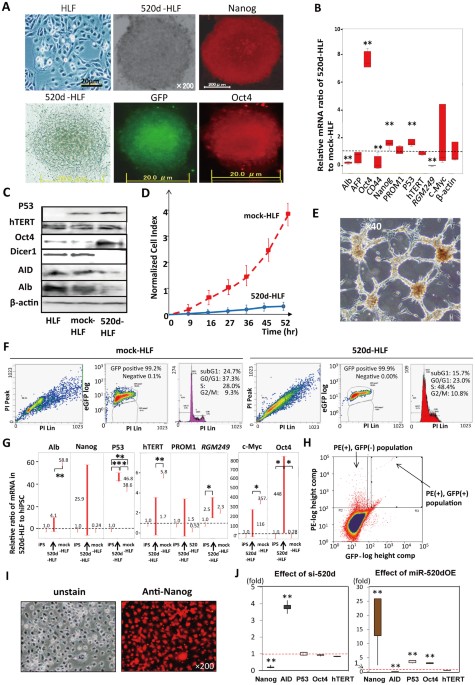
to spherical cell populations of 20–50 cells per 10-cm plate in ReproStem (Fig. 1A; top middle) and were found to express the pluripotent marker Nanog (Fig. 1A; top right). Fig. 1A shows
the morphological changes in the HLF cells (top left). Cells that were cultured in RPMI1640 expressed GFP and the pluripotent marker Oct4 (bottom). GFP was used for the identification of
transfectants by fluorescence microscopy. In all cases, the transcription of Oct4, Nanog and p53 was upregulated in 520d-HLF cells compared with mock-HLF cells at three days
post-transfection. Representative immunocytochemical findings are shown in Fig. 1A. In contrast, the _RGM249_, CD44 and albumin (Alb) expression levels were significantly downregulated (**:
P < 0.01; Fig. 1B). Western blotting showed that the Oct4 and p53 protein levels were upregulated. The expression of activation-induced cytidine deaminase (AID), Alb and Dicer1 were
suppressed in 520d-HLF cells (Fig. 1C). In migration assays, most of the pluripotent marker-positive cells could not pass through the fibronectin membranes (5 μg/ml per 6-well dish), whereas
mock-HLF cells could pass through easily (Fig. 1D). The 520d-HLF cells also had significantly lower levels of methylation markers (histone deacetylase (HDAC), Sin3A and methyl-CpG binding
domain protein 3 (MBD3)) than did the mock-HLF cells (P < 0.01; Supplementary Fig. 2C; bottom). 520d-HLF cells showed a characteristic proliferative network structure (Fig. 1E). A cell
cycle analysis of 520d-HLF cells showed increases and decreases in the S and G1/G0 phases, respectively, relative to the mock-HLF cells (Fig. 1F). hTERT (human telomerase reverse
transcriptase) and Alb were found to be downregulated in all 520d-HLF clones (n = 9) that had been cultured for more than a month in ES cell medium, compared with the levels in the HLF
cells. In contrast, the Oct4 and p53 expression levels were significantly upregulated in 520d-HLF cells and Nanog expression was elevated in eight clones. There were no significant
regulatory changes in c-Myc expression. The expression of CSC markers (PROM1) was not significantly different between HLF, hiPSCs (human induced pluripotent stem cells) and 520d-HLF cells
(Fig. 1G), but CD44 expression was significantly decreased in 520d-HLF cells (P < 0.01; Fig. 1B). From the PE-positive HLF cells, GFP (+) and alkaline phosphatase (ALP)-PE (+) cells were
selected and maintained as immature populations for 2 weeks after sorting (Fig. 1H, Supplementary Fig. 2E). The ALP-PE (+) populations strongly expressed Nanog (Fig. 1I). To confirm whether
changes in the expression of these genes provided clues regarding the underlying mechanism, we lentivirally transfected cells with a short interfering RNA (si-520d) that corresponded to
miR-520d-5p. We observed that the levels of AID and Nanog expression were the converse of those observed in cases of miR-520d-5p overexpression (520dOE). There was a significant inverse
correlation between the Nanog and AID expression levels in the si-520d and 520dOE cells. However, neither p53 nor Oct4 was downregulated in the HLF cells transfected with si-520d (Fig. 1J).
The results from the same experiments conducted in 293FT cells (rather than in HLF cells) are shown in Supplementary Fig. 2 and S1. _IN VIVO_ STUDY OF MIR-520D-VIRUS-INFECTED HLF To examine
the correlation of the _in vivo_ results with viral titer-dependent efficacies, 1.0 × 106 HLF cells were infected with 1.0 × 105 to 1.0 × 106 viral copies and athymic KSN/Slc mice were then
inoculated with the cells. Infection of cells with 1.0 × 105 (n = 8) or 3.0 × 105 (n = 22) viral copies resulted in the generation of approximately 10-mm or 1-mm undifferentiated hepatomas,
respectively, upon inoculation of mice; each mouse in the xenograft model developed a malignant tumor. Tumor volumes were significantly lower in mice that had been inoculated with cells
infected with more than 3.0 × 105 viral copies, than in mice received cells infected with 1.0 × 105 viral copies. In contrast, HLF cells that were infected with 1.0 × 106 viral copies (n =
22) did not generate any malignant tumors in mice; each recipient developed a benign tumor. Of the mice that received cells infected with 6.0 × 105 copies (n = 29), two generated homogeneous
malignant tumors, one generated a teratoma, one generated homogeneous liver tissue and one generated heterogeneous tissue that comprised both mature undifferentiated tumor cells, which were
categorized in pathological terms as a mature teratoma and normal liver tissue, which was partly accompanied by calcification (Fig. 2A). In contrast, 100% of the mock-injected mice (n = 20)
had white nodules (histologically undifferentiated hepatoma cells according to hematoxylin and eosin (HE) staining) in the peritoneum or liver (Fig. 2A; right). Only one of 20 mice that
received intraperitoneal injections generated an intraperitoneal teratoma; one mouse formed a subcutaneous liver tissue or teratoma (along the injection site) and 18 mice never developed
intraperitoneal or subcutaneous tumors or benign tissues. Heterogeneous tissues that comprised mature teratomas and liver tissues were also observed in a mouse that received cells
transfected with 6.0 × 105 copies. Thus, a viral dose-dependent suppression of tumorigenicity was observed and the ratio of malignant tumors to all tumors indicated a dose-dependent
transition to benignancy (Fig. 2B). In this study, two distinct types of benign tissues, specifically normal liver and mature teratoma-like tumors, were observed subcutaneously or
intraperitoneally. These representative tumors comprised normal liver tissue with calcification (top) and a mature teratoma with normal liver tissue (bottom; Fig. 2C). A summarized _in vivo_
study of a sufficient and effective viral titer (1 copy/cell) demonstrated that 7% and 6% of murine recipients developed normal liver tissues and mature teratomas, respectively, whereas
those that received mock-HLF cells developed undifferentiated tumors. Overall, among the 520d-HLF-injected mice, 90% (n = 64 of 71; the 71 mice included n = 22 in Fig. 2A that received 1.0 ×
106 copies) did not develop tumors, although they did generate benign or normal tissues (n = 7); each of these mice exhibited a benign tumor. Eighty-seven percent of the murine recipients
of transfected HLF cells did not form tumors or metastases. Mock-HLF recipients formed multiple peritoneal cavity nodules (Fig. 2D). A mouse that had been inoculated with 520d-HLF cells
generated intraperitoneal teratoma tissue (dermoid cyst) over an 8-week period (Fig. 2E, 2H; left, histological specimen with HE staining); during the same period, mice that had been
inoculated with mock-HLF cells generated multiple metastatic nodules that comprised undifferentiated cells (Fig. 2F). Other mice that had been inoculated with 520d-HLF generated
intraperitoneal teratomas (Fig. 2G; top, HE staining) and subcutaneous liver tissues (Fig. 2G; bottom, HE staining) over an 8-week period (size, >10 mm). The benign liver tissue specimens
included the central vein, bile ducts (Fig. 2H; right, white arrow) and hepatic cords (Fig. 2H; second from right). Moreover, in mice, 520d-HLF cells gave rise to teratomas that comprised
dermoid cysts with epidermis, sudoriferous glands, sebaceous glands (Fig. 2H; 2nd to left: white arrow) and apocrine-secreting glands over an 8-week period (Fig. 2H; top by HE staining). The
generated teratomas and liver tissues expressed GFP protein (Fig. 2I; left, HE and right, GFP, respectively). Other transfected cells did not generate any specific tissues or tumors,
including metastatic nodules. ALP and GFP-sorted 520d-HLF cells (R1, R2), which were subsequently used for metabolomic analyses and secretory protein arrays, never generated subcutaneous or
intraperitoneal tumors or other tissues in immunodeficient mice. Almost all of the hepatocytes in the liver tissues strongly expressed albumin (hAlb), as determined by immunohistochemical
staining (Supplementary Fig. S3A; left). hAlb was used as a differentiated hepatocyte marker and the _in vivo_ results indicated differentiation (Supplementary Fig. S3A; left), whereas the
_in vitro_ results revealed an undifferentiated state (Fig. 1B). α-fetoprotein (hAFP), shown in Supplementary Fig. S3A (middle) and the hepatic stellate cell/myofibroblast marker glial
fibrillary acidic protein (hGFAP), shown in Supplementary Fig. S3A (right), were weakly expressed, suggesting that 520d-HLF cells differentiated into immature liver tissues (top, 100× and
bottom, 400× magnification). Moreover, an _in vitro_ stimulation with 2 μM purmorphamine, an agent that promotes osteoblast differentiation, in RPMI1640 converted 520d-HLF cells to an
osteoblast-like phenotype (Supplementary Fig. S3B; top) and induced the significant upregulation of IBSP (osteopontin) and SPP1 expression (bone sialoprotein; P < 0.01; Supplementary Fig.
S3C). We also examined miR-520d-5p-mediated pluripotency induction in Huh7 (well-differentiated hepatoma) cells. Oct4 and Nanog expression in the small round cell population was more
elevated compared to that with mock vector induction. Four 7-day infection sessions with 1.0 × 106 copies of the viral vector per 1.0 × 106 cells were required to induce Oct4 and Nanog
expression in 520d-Huh7 cells (Supplementary Fig. S3D; top left, GFP and top right, Oct4). The Nanog, p53, Oct4, Lin28 and miR-520d-5p expression levels were upregulated, while the hAlb, AID
and _RGM249_ levels were downregulated (Supplementary Fig. S3D; bottom, see the Supplementary Figure S3D legend for an explanation of the genes examined in this study). Additionally, Oct4,
Nanog and p53 expression was upregulated and AID was downregulated in HLF cells transfected with lentiviral expression vectors (FU-tet-hOCT4, Efla_NANOG_Ires-Puro, GFP-p53 and shAID-pLKO.1);
the cells transfected with these four expression vectors grew slowly, were similar to 520d-HLF cells _in vitro_ and lost their tumorigenicity _in vivo_. METABOLOMIC ANALYSIS The following
cell lines were used in this analysis: HLF, mock-HLF, 520d-HLF (5 days after transfection (5D)), 520d-HLF (7 days after transfection (7D)) and 520d-HLF (R1 and R2), sorted with both GFP and
alkaline phosphatase (ALP). We first examined the global metabolomic profiles of induced pluripotent cells derived from cancer cells relative to the mock-HLF or 5D cells with cancerous
properties that were used as controls. We detected peaks for 165 metabolites (72 cations and 93 anions) in all groups (see Supplementary Fig. S4A for the summarized HMT analysis scheme and
Supplementary Fig. S4B and S4C for a heat map and PCA, respectively). The metabolomic profiles of 520d-HLF, R1 and R2 are very similar, which agrees with the overall genetic, epigenetic and
functional similarities that have been reported for these pluripotent cell types. We identified 165 metabolites in this study that showed a greater than two-fold difference (P < 0.01)
between the 520d-HLF (5D, 7D, R1 or R2) and mock-HLF cells (5D and 7D). Seventy-six metabolites showed a greater than 1.5-fold difference (P < 0.05) between 7D, R1 or R2 (benign cells)
and 5D (malignant cells) (Fig. 3; red or yellow columns indicate quantitative changes of greater than 2.0-fold or 1.5–2.0-fold, respectively). The data indicated high activity with regard to
nucleotide synthesis, bioenergetic production, the S-adenosyl methionine (SAM) cycle, glycolysis, the urea cycle and the production of amino acids, such as glutamine; these findings were
characteristic of cells with pluripotent or benign properties. We found that the inhibition of oxidative pathways was important to maintain pluripotency. Ketone body production and urea
cycle promotion were observed; these functions are mainly observed in the liver. Gluconeogenesis and lipid metabolism were not elevated in these transfectants, indicating that the
transformed cells might be immature and nonessential to hepatic development. Generally, the TCA cycle was not elevated in this study, although we found significant elevations in four
metabolites (malate, fumarate, succinate and cis-aconitate). Furthermore, other key metabolites were elevated, including GABA(R) and adenosyl methionine. MEASUREMENT OF
5-HYDROXYMETHYL-CYTOSINE (5-HMC; %) The general methylation levels in the transfectants were estimated and indicated slight hypermethylation in 5D and hypomethylation in R1 and R2; in
particular, there was almost no methylation in R2 (Fig. 4; top left). ALTERED GENE EXPRESSION DURING THE 520D-MEDIATED INDUCTIVE PROCESS IN HLF AFTER TRANSFECTION During the inductive
process from HLF to iPS-like cells, mediated by miR-520d-5p, the expression levels of p53, Nanog and SIRT1 were significantly upregulated (n = 4; *P < 0.05, Mann-Whitney U test),
indicating that p53, AID, Nanog and even ELAVL2 expression are correlated with benignancy and/or immaturity (pluripotency; Fig. 4). EFFECTS OF SIELAVL2 ON HLF CELLS _IN VITRO_ OR _IN VIVO_
ELAVL2 (embryonic lethal, abnormal vision, Drosophila-like 2) was predicted to be a common target candidate of miR-520d-5p, although more than 8000 genes were predicted by the databases
DIANA-MICROT, miRDB, MicroRNA.org and TargetScan VERT (Fig. 5A). Four siRNAs (short interfering RNAs) specific for ELAVL2 (siELAVL2) could induce HLF cells to form similar spheroid colonies
to those induced by miR-520d-5p (Fig. 5B). Oct4, Nanog and p53 expression were upregulated both transcriptionally (Fig. 5C) and translationally (Fig. 5D) following treatment with the four
siELAVL2, suggesting compatibility with the effects of miR-520d-5p. Immunocytochemistry revealed the upregulation of both Oct4 (upper) and Nanog (bottom) in the spheroid colonies (Fig. 5E).
We determined whether the upregulation of Oct4, Nanog and p53 and the downregulation of AID resulted in similar outcomes _in vivo_. After the regulatory protein expression levels were
confirmed in transfectants, the cells were inoculated into the right hindquarters of KSN/Slc mice (n = 8); these mice did not generate any tumors. A cell cycle analysis of HLF, pLKO.1-HLF
(mock-HLF) and siELAVL2-HLF cells (cultured in RPMI1640 or ReproStem) suggested the conversion to cell populations with homogeneous growth in phases S to G2M in the siELAVL2-HLF cells (Fig.
5F; bottom). These cells formed a network that radiated from scattered spheroid cells (Fig. 5G; top, pLKO.1-HLF as a mock control and bottom, siELAVL2-HLF). A luciferase reporter expression
assay demonstrated the suppression of ELAVL2 gene expression by the binding of miR-520d-5p to the 3'UTR of ELAVL2. The ELAVL2 3'UTR sequence (1356–3805) contains 10 putative
binding sites for miR-520d-5p (Supplementary Fig. S5) and miR-520d-5p bound to two of the most potent sites (508–525 and 880–894 in the 3'UTR and 1853–1880 and 2235–2249 of ELAVL2 gene)
in the ELAVL2 3'UTR, resulting in the inhibition of ELAVL2 translation relative to the controls miR-520d-3p or pLKO.1 (contains a mismatch sequence within miR-520d-5p (see Methods);
Fig. 5H). Both synthesized miR-520d-5p and the miR-520d-5p expression vector could specifically reduce ELAVL2 translation by approximately 50–70%, compared to controls transfected with
synthesized miR-520d-3p or pLKO.1. A control expression vector that included mismatch sequences within miR-520d-5p did not significantly affect the luciferase expression, which was similar
to that of the empty pLKO.1 vector. An _in vivo_ study in mice inoculated with ReproStem-cultured siELAVL2-HLF cells did not result in the formation of tumors, liver tissues or scars (Fig.
5I). Xenografted mice, which received siELAVL2-HLF cells that had been maintained with RPMI1640, generated undifferentiated tumors (2/16), no tumors (4/16), scars (8/16; top in Figure 5I)
and differentiated tumors (2/16). The individual cells had swollen and clear nucleoli and cytoplasm with weak basophilia, a vacuole and a fat drop. Many neoplastic cells resembled
fibroblasts or myofibroblasts and the presence of a blood vessel system supported the possibility of differentiation. The immature differentiated tumors revealed the possible presence of
fibroblasts, myofibroblastic forms, fat drops, collagen fibers, cytoplasmic vacuoles and/or an immature blood vessel system (Fig. 5I; bottom right, 200× HE staining). However, because ELAVL2
expression increased toward the mock level in siELAVL2-HLF, Huh7 or 293FT cells, presumably it was not needed to maintain the undifferentiated status. Additionally, a mouse that received
HLF cells with upregulated Nanog and downregulated AID expression generated a small tumor that comprised immature and differentiated regions such as collagen fibers or cytoplasmic vacuoles
(1/16). MIR-520D-RELATED DEDIFFERENTIATION INDUCTION IN HUH7 CELLS _IN VIVO_ Once per week, we infected cells with viral constructs _in vitro_. All mock-Huh7 recipients formed
well-differentiated hepatomas (Supplementary Fig. S3E; n = 10), but 50% (n = 4) of the mice inoculated with 520d-Huh7 cells after a 1-week transfection generated less-differentiated tumors
after a month (Supplementary Fig. S3F; left); the remaining mice did not generate tumors. HE staining (200× magnification) revealed poorly differentiated hepatomas (Supplementary Fig. S3G;
left) and poorly-differentiated hepatoma cells (Supplementary Fig. S3G; right). Mice that received the transfection for one month did not generate tumors after one month (n = 4), indicating
the induction of dedifferentiation in a dose-dependent manner; however, more time was required for the loss of tumorigenicity in the Huh7 recipients. Additionally, we examined the
tumorigenicity of HLF cells with upregulated Nanog and downregulated AID expression (Fig. 1J; n = 16); these cells led to no tumor formation (n = 15) or small undifferentiated subcutaneous
tumors with an immature and differentiated tumor including collagen fibers or cytoplasmic vacuoles, unlike the undifferentiated hepatoma cells. The pathological findings were similar to
those of HLF cells treated with siELAVL2 (n = 2; Fig. 5I; bottom). Subsequently, we examined the tumorigenicity of HLF cells with upregulated Oct4, Nanog and p53 and downregulated AID
expression in athymic mice; these cells yielded no tumor formation (n = 8). The observed phenotypic changes in HLF cells that were infected with four lentiviral vectors were similar to those
observed in 520d-HLF cells _in vitro_. EFFECT OF MIR-520D-5P ON HUMAN FIBROBLASTS NHDF-Neo (left) and Ad (right; Supplementary Fig. S6A; 200× magnification) were infected with a
miR-520d-5p-expressing lentiviral vector, as described previously. The transfectants showed no phenotypic changes when compared with parental cells, but they had a longer lifespan (more than
three months) until senescence induction than did the parental cells. Immunocytochemistry revealed upregulated Nanog (right) or Oct4 expression (middle) in the miR-520d-5p-expressing
fibroblasts (520d-NHDF-Neo and 520d-NHDF-Ad in each; Supplementary Fig. S6B, S6C). To examine the tumorigenicity of the 520d-NHDF-Neo or 520d-NHDF-Ad cells in KSN/Slc mice, transfectants
were inoculated into the right hindquarters of KSN/Slc mice. Neither the 520d-NHDF-Neo (n = 8) nor the Ad (n = 8; right) cells generated tumors in mice at 12 weeks post-inoculation
(Supplementary Fig. S6D). DISCUSSION miRNAs have been implicated in numerous biological processes16. Accumulating data suggest that miRNA dysregulation occurs frequently in various
carcinomas, including those of the lung, colon, stomach, pancreas and liver3,17. Many recent studies have attempted to better understand the roles of miRNAs and reveal the function and
importance of miRNA-mediated regulation in cancer and normal cells18. Aberrant miRNA expression profiles that correlate with particular tumor phenotypes can also be used to distinguish
between normal tissues and tumors19. Perturbations of miRNA expression levels might also lead to anti-tumorigenesis20. Thus, the dual effects of miRNAs in carcinogenesis and normal stem cell
differentiation strongly suggest that miRNAs might be involved in the transformation of normal stem cells to CSCs. Although our current understanding of miRNA functions remains incomplete,
this study has helped elucidate the intricate roles of miRNAs in the regulation of cellular processes such as differentiation, reprogramming and oncogenesis. This is the first study to
provide evidence that a single miRNA can convert malignant or immortalized cells to benign or normal cells. The stemness-mediated process does not necessarily result in normal cells, but
normal hiPSCs form teratomas _in vivo_21,22,23 and it was thought that tumorigenicity might cause methylation in cells prior to developmental growth24,25,26. In this study, western blotting
and RT-PCR were used to show that p53 was stably upregulated in both miR-520d-5p-lentivirus-infected 293FT cells (520d-293FT; hsa-miR-520d-5p-overexpressing 293FT) and 520d-HLF cells, unlike
the p53 levels in hiPSCs (Figure 1, Supplementary Fig. S2). High titers (600,000–1,000,000 copies) of the miR-520d-5p-expressing lentiviral vector increased the efficacy of this induction
through stable changes in translational regulation (Figure 2). Compared to the mock virus-infected 293FT (mock-293FT) and mock-HLF cells, the DNA contents of the 520d-293FT and 520d-HLF
cells were increased in the subG1 and S phases, respectively (Figure 1F, Supplementary Fig. S1A). A comparative analysis of the DNA methylation patterns in 520d-293FT and 520d-HLF cells
showed the reverse pattern (Supplementary Fig. S1C), presumably due to differences in the cell differentiation levels. AID, which binds to the promoter regions of Nanog or Oct4 and regulates
pluripotency, was downregulated in both cell types, compared to the parental cells. However, in culture conditions intended to maintain undifferentiated cells, post-transfection AID
expression was upregulated in 520d-293FT and downregulated in 520d-HLF cells, indicating the AID-independent regulation of pluripotent markers (Oct4 and Nanog), as these proteins were
upregulated in both cell types27. Many cancer cells also express Oct4 and Nanog and there are many similarities between cancer cells and embryonic stem cells in terms of the stemness
process28. However, many types of cancer cells downregulate p53 and cannot generate teratomas or original tissues _in vivo_. DNMT1 (DNA (cytosine-5-)-methyltransferase), which is upregulated
in preneoplastic lesions and poorly differentiated tumors, was also downregulated29 and a compensatory HDAC dysregulation was possibly induced because DNMT1 could not bind to HDAC. Sin3A
upregulation might repress Myc and thus exert an anti-oncogenic effect on 293FT cells30. The crucial step toward normal methylation or epigenetic modification might be the suppressive
regulation of AID and DNMT1, as both proteins play important roles in carcinogenesis and cancer progression24,25,26,27. It is possible that either a DNMT or an AID inhibitor alone could
reprogram mouse fibroblasts to iPSCs without viral induction, but this has not yet been achieved and we think that AID might be a crucial factor in the reprogramming of cancer cells such as
KLM-1, PK9, PK-45p or HT1080, as well as HLF, because AID expression might negatively regulate Oct4, Nanog and p53 (Figure 6). miR-520d-5p converted even well-differentiated hepatoma cells
(Huh7) to non-tumorigenic cells via less-differentiated and tumorigenic hepatoma cells. A stringent conditioning regimen directed toward differentiation should be determined because the
histology of the generated tissues was almost homogeneous and the outcomes of the diverse phenotypes (teratoma, hepatic tissue or no tumor formation) _in vivo_ might depend on the
reprogramming level and/or p53 expression levels. Additionally, the metastatic properties of 520d-HLF were suppressed _in vitro_ and _in vivo_. The DNA percentage of 520d-HLF cells was
increased in the S phase; this result might be compatible with a report in which mir-302a/b/c/d coexpression drove homogeneous proliferation without apoptosis and enhanced the G1 phase
pathways31. This unexpected conversion to normal liver tissues implies therapeutic potential against well-differentiated and poorly differentiated hepatomas. Similarly, the regulation of
malignancy by miR-520d-5p in other cancers appears not to be caused by a loss of metastatic properties but by the conversion of cancer cells to normal stem cells through a dedifferentiation
process and the maintenance of upregulated p53 levels; this has not been observed previously in hiPSCs (Fig. 4)32. Differences between the normal liver tissues and benign teratomas (dermoid
cysts) might depend on the cellular miR-520d-5p or p53 expression levels. _In vivo_, the upregulation of Oct4, p53 and _RGM249_ and the downregulation of c-Myc, hTERT and Alb expression were
characteristic genetic changes. The PROM1 expression levels did not change significantly33, but the downregulation of CD4434 and upregulation of CD90, CD105 and CD13, which are mesenchymal
stem cell markers, were significant at the transcriptional level (P < 0.01)35. This study further indicates that the miR-520d-5p overexpression in HLF cells downregulated Dicer1
expression, although the opposite was observed in 293FT cells. Major regulators of miRNA biogenesis (e.g., Dicer and Drosha) are downregulated in cancer cells36, suggesting that the miRNA
regulatory mechanisms and requirements might be different in cancer cells and 293FT cells and these differences might be required to reprogram cancer cells and suppress miRNA biogenesis. The
optimal conditions for the induction of each phenotype from HLF _in vivo_ remain undetermined, although these inductions are assumed to be associated with reprogramming states or the
homogeneity of the cellular states37. In the _in vivo_ study, infection of HLF cells with a sufficient lentiviral titer induced the generation of homogeneous benign or normal tissues,
whereas HLF cells infected with virus at an insufficient titer induced the generation of heterogeneous tissues that comprised two populations (normal liver tissue and mature teratoma); we
therefore concluded that lentiviral integration did not promote the differentiation of transfectants into diverse phenotypes and that sufficient transfection efficiencies would reproducibly
induce the transition of malignant cells to benign or normal states. _In vitro_, 520d-HLF differentiated into osteoblasts38, suggesting differentiation into mesodermal tissue. Additionally,
these cells demonstrated differentiation into endodermal (liver tissue) and ectodermal (epidermis) tissues _in vivo_, indicating that 520d-HLF the homogeneously converted to hPSCs, not CSCs.
Surprisingly, undifferentiated cancer cells with strong metastatic and chemotherapy-resistant properties also converted to normal liver tissues. In a preliminary study of other types of
undifferentiated malignant cells (glioblastoma multiforme, anaplastic thyroid cancer, pancreatic cancer and fibrosarcoma cells), we confirmed no tumor formation, the loss of malignant
properties and ductal cell carcinoma conversion to pancreatic acinar cells or lipid-producing cells despite a presumably similar mechanism as that involved in the miR-520d-5p-induced
conversion of HLF (data not yet available). These malignancies might be treated easily with miR-520d-5p, although they are known clinically to be extremely intractable malignancies.
miR-520d-5p induction might be more sensitive in CSC because undifferentiated malignancies should contain CSCs. Although Nanog or Oct4 can bind to the DNMT1 promoter region to drive
transcriptional upregulation39, DNMT1 expression was downregulated during this conversion in the current study. Methylation or demethylation might not be crucial events in the maintenance of
pluripotency or the upregulation of p53. Although AID upregulation induces p53 mutations in somatic cells, leading to p53 downregulation and subsequent carcinogenesis40, AID downregulation
might play a part in p53 upregulation; however, the mechanism of p53 upregulation remains unclear. Unlike miR-520d-5p, siAID alone was not able to induce a phenotypic change. Gene-specific
hypermethylation might silence developmentally related genes, leading to dedifferentiation41. The measurement of 5-hydroxymethyl-cytosine (5-hmC) revealed a more hypomethylated status in 7D,
R1 and R2 and slight hypermethylation in 5D (Fig. 4)42. Therefore, we propose a mechanism for how this miRNA induces the upregulation of both Nanog and Oct4 in normal cells and cancer
cells, as shown in Fig. 6. In a previous report, a comparative metabolomic analysis of ESCs and iPSCs revealed similar differences in the levels of metabolites that are involved in
transmethylation, cellular respiration and energy production; however, the S adenosylmethionine (SAM) cycle includes 5′-methylthioadenosine, hypoxanthine and inosine, which drive the
methylation process and all of these were significantly elevated in iPSCs compared with ESCs. Additionally, adenosyl methionine decarboxylase (AMD1) plays a crucial role in the renewal of
normal stem cell pluripotency43. The methyl group belonging to SAM is the most common substrate in transmethylation reactions such as DNA methylation15. Additionally, the fatty acid (ω-3 and
ω-6) levels were elevated in ESCs. In other words, the unsaturated fatty lipid levels were lower in iPSCs than in ESCs. In somatic cells, the oxidative: glycolytic energy production ratios
were close to those in iPSCs. However, the oxidative: glycolytic energy production ratios in 520d-transfectants were lower than those in iPSCs. According to a comparative metabolic analysis
of laser-dissected normal tissues and colon or gastric cancer tissues, the TCA cycle metabolites were not significantly elevated in the normal tissues44. Additionally, increased levels of
citric acids in the TCA cycle lead to the suppression of glycolysis, but the citric acid levels were not found to be significantly increased in this study, suggesting that glycolysis was
occurring. Thus, comparative metabolic profiling of the transfectants with malignant properties and with normal or benign properties showed that glycolysis, the urea cycle and amino acid
synthesis were promoted, indicating the possibility that HLF cells might be converted to cells with stemness and immature somatic properties because the profile results were compatible with
those of previous reports. Given that the bioenergetics of various somatic cells correlated with their reprogramming efficiencies15, these findings might be applicable to cancer cells
derived from somatic cells; more immature hepatomas might revert more easily to normal pluripotent cells with somatic properties because more immature cancers have higher bioenergetic
levels. However, changes in the activities of different pathways such as glycolysis, nucleotide synthesis and the urea cycle cannot be ascertained from differences in the total metabolite
statuses between different cell types alone and therefore the use of stable isotope tracers, coupled with metabolomics analysis, must be conducted to warrant conclusions on metabolic pathway
activity in future studies. This is because a given metabolite, such as lactate or ATP, can be synthesized from or consumed by a number of different pathways. Without isotope tracing, it
would not be possible to confirm that lactate accumulation comes from glycolysis or that ATP buildup results from de novo nucleotide synthesis. Moreover, the majority of lactate produced
during glycolysis is expected to be released into the medium, which was not analyzed in this report; also, the total ATP level is most likely sustained by oxidative or substrate-level
phosphorylation (not measured in this report). Therefore, the present data only showed a tendency with regard to the difference between the malignant properties and benign properties of
520d-HLF cells. However, there might be a meaningful relationship between the overall reduced metabolism in 5D cells and the progression of demethylation (Figure 3, 4 & Supplementary
Fig. S3H). ELAVL2 (HuB) might be a target gene of miR-520d-5p. ELAVL2 is predicted to contain ten binding sites for miR-520d-5p (Supplementary Fig. S5) in its 3'UTR sequence
(1356–3805), suggesting that miR-520d-5p and siELAVL2 might bind at least two sites in the 3'UTR. ELAVL2 might play a crucial role in carcinogenesis or cancer stemness because
transfectants (by miR-520d-5p or siELAVL2) that were cultured in ReproStem for more than two weeks did not generate tumors. The upregulation of p53, as well as Oct4 and Nanog, was induced by
miR-520d-5p and siELAVL2 in an AID- or DNMT1-independent manner. Additionally, a month was required (four times at 1.0 × 106 copies/week) to convert well-differentiated hepatoma cells
(Huh7) to non-tumor forming cells, suggesting that miR-520d-5p might drive transfectants strongly toward dedifferentiation. Additionally, 520d-5p overexpression in fibroblasts (both NHDF-Ad
and Neo) did not induce the formation of spheroid pluripotent cells _in vitro_ or promote the acquisition of tumorigenicity in these cells (n = 6 for each; Supplementary Fig. S6D). In
conclusion, herein, we did not elucidate the target gene of miR-520d-5p. However, we report a noteworthy function of hsa-miR-520d-5p and describe presumably key roles for p53, AID, Nanog and
the miR-520d-5p target genes, including ELAVL2, TEAD1, SBF2, PUM2, GATAD2B and SEH1L, in the benign conversion process (Fig. 5), in which well-differentiated and undifferentiated hepatoma
cells were converted to normal/benign phenotypes via stemness induction, resulting in a reversal of malignancy _in vivo_. At present, we cannot explain these phenomena with regard to gene
expression and metabolite production. Hence, this report contains speculations derived from raw data. Although the notion that cancerous cells cannot revert to their original, healthy,
states is widely accepted, this idea might no longer be valid. METHODS CELLS To determine the _in vitro_ and _in vivo_ effects of hsa-miR-520d-5p expression, we used two cell lines and
lentiviral vectors. hiPSCs (HPS0001 and HPS0002) and hepatoma cells (HLF and Huh7) were provided by the RIKEN BioResource Center Cell Bank45 and the Cell Resource Center for Biomedical
Research, Institute of Development, Aging and Cancer Tohoku University, respectively. Poorly differentiated or undifferentiated (HLF) and well-differentiated (Huh7) human hepatoma cell lines
with strong and weak _RGM249_ expression, respectively, were cultured in RPMI1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin. To maintain the undifferentiated states of
virus-transformed cells, the cells were cultured in ReproStem medium (ReproCell, Tokyo, Japan) with 10 ng/ml of bFGF-2. To examine the effects of miR-520d-5p on normal human dermal
fibroblasts _in vitro_ and _in vivo_, we used adult NHDF (NHDF-Ad) and neonatal NHDF (NHDF-neo) cells (TAKARA BIO, Tokyo, Japan) and cultured them with the FGM-CD Bullet Kit (TAKARA BIO).
Additionally, for comparison with HLF cells or hiPSCs, the human mesangial cell line 293FT (Invitrogen Japan K.K., Tokyo, Japan) was used for its potent facility for hiPSC induction, as
previously reported46. 293FT cells were cultured in DMEM supplemented with 10% FBS, 0.1 mM MEM non-essential amino acid solution, 2 mM L-glutamine and 1% penicillin/streptomycin. LENTIVIRAL
VECTOR CONSTRUCT To examine the effects of miR-520d-5p overexpression, we transfected pMIRNA1-miR-520d-5p/GFP (20 μg; System Biosciences, Mountain View, CA, USA) or the mock vector pCDH (20
μg) into 293FT and HLF cells (5 × 106 cells/10 cm culture dish). To investigate the effects of miR-520d-5p silencing, we transfected pRNATinH1.4/Lenti-520d-5p (20 μg; Genscript, Piscataway,
NJ, USA) or the mock vector pRNATinH1.4/Lenti (20 μg) into HLF cells. To harvest viral particles, the cells were centrifuged at 170,000 × _g_ (120 min, 4°C). The viral pellets were collected
and viral copy numbers were measured with a Lenti-X™ qRT-PCR Titration kit (Clontech, Mountain View, CA, USA). For 293FT or HLF cell infection, one million lentiviral copies were used per
10-cm culture dish. We transfected 50 nM synthesized oligonucleotides into 293FT cells with FuGENE HD Transfection Reagent (Roche Diagnostics, Basel, Switzerland). To confirm the induction
of 520d-HLF differentiation into osteoblasts, the cells were treated with 2 M purmorphamine in routine RPMI1640 medium for one week. To confirm the status of ELAVL2 as a candidate target
gene of miR-520d-5p, four siRNAs (AAD63-B9, 10, 11 and 12) specific for ELAVL2 (shELAVL2-pLKO.1) were purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). pLKO.1 and scramble
siRNAs were used as controls. The siRNAs were transfected together into HLF cells via lentiviral constructs according to the optimized method. The representative gene expression levels in
HLF cells that were treated with si-520d or 520dOE via pRNATinH1.4/Lenti or pMIRNA/Lenti/GFP, respectively, are shown. To estimate the effects of doxycycline-treated si-520d or 520dOE,
doxycycline-untreated HLF cells or pCDH/lenti/GFP-treated cells were used as controls. To confirm the effects of Oct4, Nanog and p53 upregulation and AID downregulation in HLF cells,
lentiviral vectors (FU-tet-hOCT4, Efla_NANOG_Ires-Puro and GFP-p53) were purchased from Addgene (Cambridge, MA, USA), while shAID-pLKO.1 was purchased from Thermo Fisher Scientific Inc.
(Waltham, MA, USA). The ELAVL2-specific shRNA was purchased from Genecopoeia (Rockville, MD, USA). The siRNA sequences for ELAVL2 were as follows: B9, 5′-ccgugacuuuaacaccautaa-3′; B10,
5′-cgcauuauuacuucucguauu-3′; B11, 5′-gucuccuuuaagacaaacaaa-3′ and B12, 5′-gcuucuaucagagaugcaaau-3′. The scrambled sequences for miR-520d-5p and ELAVL2 were 5′-gaguccgcctcuatagacaa-3′ and
5′-auucguuguuauugu-3′, respectively. IMMUNODEFICIENT MICE AND _IN VIVO_ EXPERIMENTS After a 1-week lentiviral infection, 5 × 107 HLF cells were harvested and injected intraperitoneally or
subcutaneously into the right flank (injection volume, 200 μl) of each mouse. Six-week-old immunodeficient mice (KSN/Slc; Shimizu Laboratory Supplies, Kyoto, Japan) were fasted for 24 hours
before the experiments but were allowed free access to water and feed for 4–12 weeks after the injections; subcutaneously injected mice (n = 51) were observed for 4–8 weeks, while
intraperitoneally injected mice (n = 20) were observed for 8 weeks prior to anesthetization with 100 mg/kg Nembutal and then sacrificed for anatomic and histological examinations. Prior to
the endpoint, 59 mice were inoculated subcutaneously in a dose-dependent manner (8 mice for a 10% transduction efficiency, 22 for 30% and 29 for 60%). Next, 20 mice received 520d-HLF cells
that had been sorted for ALP and GFP positivity to examine the influence of differences in culture conditions between media that maintained the undifferentiated status and RPMI1640/10% FBS.
Tumor volumes were determined according to the following formula: volume = π/6 × width × length × height. To examine the conversion of well-differentiated hepatoma cells (Huh7) to
pluripotent cells with concomitant p53 upregulation or examine the effects of miR-520d-5p on human fibroblast cell lines, an _in vivo_ study was performed in immunodeficient mice. After
transfection with the same viral vehicle titer used in the previous _in vivo_ study with HLF cells, the tumorigenicities of the transfectants (520d-5p-Huh7, 520d-5p-NHDF-neo and
520d-5p-NHDF-ad) were confirmed at one week or one month after inoculation for 520d-5p-Huh7 or at three months for 520d-5p-NHDFs. All animals were housed and fed in the Division of
Laboratory Animal Science of Tottori University under a protocol that was approved by the Japanese Association for Accreditation for Laboratory Animal Care and the animal research and
handling were performed in strict conformance with federal Institutional Animal Care and Use Committee guidelines. All experiments reported in this study were approved by an institutional
committee. RT-PCR Total RNA, inclusive of the small RNA fraction, was extracted from cultured cells or homogenized mouse tissues with the mirVana miRNA Isolation Kit (Ambion, Austin, TX,
USA). Mature miRNAs were quantified with a Mir-X™ miRNA qRT-PCR SYBR kit (Clontech) according to the manufacturer's instructions. miRNA (25 ng/μl) was quantified with a Mir-X miRNA
qRT-PCR SYBR Kit (Takara Bio Inc., Tokyo, Japan) to confirm siRNA suppression and evaluate changes in miRNA expression. Gels were run under the same experimental conditions. PCR and data
collection analyses were performed with a BioFlux LineGene (Toyobo, Nagoya, Japan). The expression levels in the samples were determined according to the standard curve method. All data,
except those for hTERT, were normalized to the internal control β-actin. hTERT expression was estimated from the copy number, according to a previously developed quantification method47. U6
small nuclear RNA was used as an internal control. Total RNA (50 ng/μl) was reverse transcribed and amplified with the OneStep RT-PCR kit (Qiagen, Tokyo, Japan). RNA quantification was
confirmed by high-reproducibility sequencing. Supplementary Table S1 shows the primer sequences that were used for mRNA or miRNA quantification. The data were analyzed statistically with a
one-way ANOVA or Mann-Whitney U test and significant differences are shown as *: P < 0.05 and **: P < 0.01. WESTERN BLOTTING We performed western blotting with 20 μg/μl of proteins and
the i-Blot gel transfer system (Invitrogen, Tokyo, Japan). Anti-hTERT, p53, Oct4, DICER1, AID, albumin (Alb) and ELAVL2 antibodies were used at 1:500 dilutions and the anti-β-actin antibody
was diluted 1:1,000 according to the manufacturer's instructions. Chemiluminescent signals were detected within 1 min with LAS-4000 (Fujifilm, Tokyo, Japan). The gels were run under
the same experimental conditions. MIGRATION ASSAY The invasive abilities of transfectants were estimated with a CIM-Plate 16, which detects cell invasion/migration in real time, according to
the manufacturer's instructions (xCELLigence system, Roche, Basel, Switzerland). IMMUNOCYTOCHEMISTRY The immunohistochemical examination was performed with antibodies to detect
pluripotent markers (anti-Oct4 or anti-Nanog) and with an Embryonic Stem Cell Marker Antibody Panel, according to the manufacturer's instructions (R&D Systems, Minneapolis, MN,
USA). 293FT, Huh7 and HLF cells were infected with lentiviral particles that contained hsa-miR-520d-5p. Floating transfectants were harvested and transferred to new culture dishes for
microscopic examinations or to slide chambers for immunostaining. IMMUNOHISTOCHEMISTRY During the immunohistochemical analyses, 4% paraformaldehyde-fixed liver tissue specimens were
processed48. The following monoclonal antibodies were used: anti-hAlb, anti-hAFP and anti-hGFAP (Sigma, St Louis, MO, USA). As a negative control, tissues were stained without the primary
antibody. A pathologist estimated the degree of protein expression. CELL CYCLE AND CELL SORTING ANALYSIS Cell cycle analysis and cell sorting were conducted to confirm that the 520d-293FT
and 520d-HLF cells comprised pluripotent cell populations. The DNA content was analyzed with a flow cytometer (Epics Altra; Beckman Coulter Inc., CA, USA); the cells were assessed from
approximately 20,000 collected events after the transfection of a pMIRNA1-miR-520d-5p/GFP clone, using EXPO32 ADC Analysis software. GFP-positive cells were sorted on a Moflo XDP cell sorter
(Epics Altra; Beckman Coulter Inc.). In detail, for cell cycle analysis, a single cell suspension was washed once with cold PBS. The cell pellet was loosened by shaking the tube gently and
was fixed with 3.7% formalin in ddH2O, added dropwise. The cells were then incubated at least overnight at −20°C. After fixation, the cells were washed twice with cold PBS to remove the
EtOH, resuspended at 1 × 106 cells/ml in PBS with 100 U/ml RNaseA and incubated for 50 min at 37°C. Next, 50 μg/ml of propidium iodide were added and the mixture was incubated for 40 min on
ice in the dark. The DNA content was analyzed with a flow cytometer; the cells were assessed from approximately 20,000 collected events after the transfection of a pMIRNA1-miR-520d-5p/GFP
clone, using the EXPO32 ADC Analysis software. GFP-positive cells were sorted on a Moflo XDP cell sorter. Next, the flow cytometric purification of GFP-expressing or PE-positive cells was
performed. The cells were maintained in an immature state for two weeks after viral transduction. 293FT and HLF cells were resuspended in PBS with 5% FCS. After the cells were stained with a
PE-conjugated anti-ALP antibody, GFP or PE-positive cells were sorted and analyzed on a Moflo XDP cell sorter against an argon laser (488 nm, 100 mW). A total of 1 × 108 cells were analyzed
for forward scatter, side scatter and PE and GFP fluorescence. The FL1 and FL2 channels were used to detect GFP and PE, respectively. HISTOLOGICAL EXAMINATION Lung, liver, intraperitoneal
or postperitoneal metastases and tumor volumes were investigated macroscopically or under a dissecting microscope with bright-field imaging. Tissue samples were fixed in 10% buffered
formalin overnight, washed with PBS, transferred to 70% ethanol, embedded in paraffin, sectioned and stained with hematoxylin and eosin (HE). FLUORESCENCE DETECTION IN CELLS To estimate the
efficacy of infection with the miR-520d-5p-expressing lentiviral vector, GFP expression was detected with an OLYMPUS IX71 microscope and TH4-100 power supply for the microscope (Tokyo,
Japan). TARGET PREDICTION OF SPECIFIC MIRNA Algorithms were used to predict the specific miRNA targets (Cosmo Bio, Tokyo, Japan). Fig. 5A shows the predicted target genes of hsa-miR-520d-5p.
For the Venn diagram, we used the following databases: TargetScanHman6.0 (http://www.targetscan.org/vert_60/), microRNA.org (http://www.microrna.org/microrna/getMirnaForm.do), Pictar
(http://pictar.mdc-berlin.de/), RNA hybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) and miRBase (http://www.mirbase.org/). METABOLOMIC ANALYSIS AND SAMPLE
PREPARATION To examine the metabolic profiles associated with induced pluripotency accompanied by non-cancerous properties, we used an untargeted metabolomics approach to analyze the
relative abundances of metabolites in normal or pluripotent cells derived from human hepatoma cells (HLF). The quantified measurements and analysis of approximately 900 metabolites were
performed by Human Metabolome Technologies (HMT; Tsuruoka, Yamagata, Japan). The following cell types were used in this study: HLF, mock-HLF, 520d-HLF (5D), 520d-HLF (7D) and 520d-HLF (R1
and R2) sorted with both GFP and alkaline phosphatase (ALP). In detail, the five days post-transfection group (5D) presented with malignant tumors _in vivo_, whereas 7D, R1 (cells sorted
with pluripotent markers and GFP that led to teratoma formation or normal liver tissue development) and R2 (cells sorted with pluripotent markers and GFP that led to no tumor development)
presented with benign phenotypes _in vivo_. CE-TOFMS was conducted using an Agilent Capillary Electrophoresis System equipped with an Agilent 6210 Time of Flight mass spectrometer, an
Agilent 1100 isocratic HPLC pump, an Agilent G1603A CE-MS adaptor kit and an Agilent G1607A CE-ESI-MS sprayer kit (Agilent Technologies, Waldbronn, Germany). LC-TOFMS was conducted on an
Agilent 1200 HPLC pump equipped with an Agilent 6210 Time of Flight mass spectrometer (Agilent Technologies). These systems were controlled by the Agilent G2201AA ChemStation, version
B.03.01, for CE and MassHunter for LC software packages (Agilent Technologies). Data acquisition was performed with Analyst QS Build: 7222 software for the Agilent TOF (Applied Biosystems,
Foster City, CA, USA/MDS Sciex, Ontario, Canada). Measurements of the extracted metabolites were performed using CE-TOFMS as described previously49 with a commercial electrophoresis buffer
(Solution ID H3302-1021; Human Metabolome Technologies Inc., Tsuruoka, Japan). Peak extraction was conducted with a modified version of the open source software MathDAMP50. Peak alignment
was performed according to the m/z values and normalized migration times. Next, the peak areas were normalized against those of the internal standards MetSul and CSA for cationic and anionic
metabolites, respectively. The resulting relative area values were further normalized to the sample amount (105 cells) and the average values were calculated for each group. Annotation
tables were produced from the CE-TOFMS and LC-TOFMS measurements of standard compounds and aligned with the datasets according to similar m/z values and shifts in the retention times of the
LC-TOFMS measurements. Student's _t_ test was used for comparisons of compound levels between sample groups. The metabolic pathway map used was the Analysis of Networks containing
Experimental Data (VANTED, http://vanted.ipk-gatersleben.de/)51. To examine the metabolic profile associated with induced pluripotency accompanied by non-cancerous properties, we used an
untargeted metabolomics approach to analyze the relative abundances of metabolites in normal or pluripotent cells derived from HLF. MEASUREMENT OF 5-HYDROXYMETHYL-CYTOSINE (5-HMC; %)
5-Hydroxymethlyl-cytosine was measured in HLF, mock-HLF and 520d-HLF cells (3D, 5D, 7D, R1 and R2; 200 ng each) with the MethylFlash Hydroxymethylated DNA Quantification kit (Colorimetric)
according to the manufacturer's instructions (EPIGENTEK, Farmingdale, NY, USA). ESTIMATION OF ELAVL2 AS A TARGET GENE OF MIR-520D-5P Based on the predicted target genes of miR-520d-5p
(MIMAT0002855: cuacaaagggaagcccuuuc) according to various databases (miRBase; http://www.mirbase.org, DIANA-MICROT; http://diana.cslab.ece.ntua.gr/DianaTools/, miRDB; http://mirdb.org,
RNA22-HAS; http://cm.jefferson.edu/rna22v1.0, TargetMiner; http://www.isical.ac.in/~bioinfo_miu, mircoRNA.org; http://www.microrna.org/microrna and TargetScan-VERT;
http://www.targetscan.org/cgi-bin/targetscan/vert_50) and after confirming gene downregulation by RT-PCR, we examined gene expression in cells transfected with four types of siRNAs against
ELAVL2 (siELAVL2-HLF; see the lentiviral vector construct section) and compared this with gene expression in 520d-HLF cells. We performed RT-PCR, western blotting, immunocytochemistry and
cell cycle analysis as described previously. To investigate the binding of miR-520d-5p to the 3'UTR of ELAVL2, sequences (or mock mismatch sequences) that corresponded to predicted
sites were ligated into the multiple cloning site (MCS; PmeI and XhoI) of psiCHECK-2 (Promega KK, Tokyo, Japan). The signals were detected with a dual luciferase reporter expression assay
(Promega KK, Tokyo, Japan). Synthesized miR-520d-5p (MBL, Nagoya, Japan) or pMIR-520d-5p were co-transfected into HLF cells with each prepared expression vector. Synthesized hsa-miR-520-3p
(MBL, Nagoya, Japan) or pLKO.1 (Addgene, Cambridge, MA, USA) with the mismatch sequence were used as controls. Forty-eight hours after transfection, the luciferin expression (RLU) was
measured on an Infinite F500 (TECAN, Männedorf, Switzerland). RLU (renilla/firefly) activity was standardized to that of a control (n = 4). siELAVL2-HLF cells were injected into the right
hindquarters of immunodeficient mice that were fed routinely for two months and observed for tumorigenicity and estimations of tumor quality. The following four pairs of sequences were
inserted into psiCHECK-2: ELAVL2-3'UTR sense 1, 5′-TCGAGAACAGTATTTATTTTGTAAGTTT-3′; ELAVL2-3'UTR antisense 1, 5′-AAACTTACAAAATAAATACTGTTC-3′; ELAVL2-3'UTR sense mock 1,
5′-TCGAGAACAGTATTaaTATTTTGTAAGTTT-3′; ELAVL2-3'UTR antisense mock 1, 5′-AAACTTACAAAATAttAATACTGTTC-3′; ELAVL2-3'UTR sense 2, 5′-TCGAGACGTCCTGCTTTTTGTAGTTT-3′; ELAVL2-3'UTR
antisense 2, 5′- AAACTACAAAAAGCAGGACGTC-3′; ELAVL2-3'UTR sense mock 2, 5′- TCGAGACGTCCTGCaaTTTTTGTAGTTT-3′; and ELAVL2-3'UTR antisense mock 2, 5′-AAACTACAAAAAttGCAGGACGTC-3′.
STATISTICAL ANALYSIS A Mann-Whitney U test or one-way ANOVA was used for comparisons between the control, mock and miR-520d-5p or si-520d groups with one observed variable. P < 0.05 was
considered significant (*: P < 0.05, **: P < 0.01). In the box plots, the top and bottom of each box represent the twenty-fifth and seventy-fifth percentiles, respectively, thus
providing an interquartile range. The line through the box indicates the median and the error bars indicate the fifth and ninety-fifth percentiles. REFERENCES * Lee, Y. et al. MicroRNA
maturation: stepwise processing and subcellular localization. EMBO J. 21, 4663–4670 (2002). Article CAS PubMed PubMed Central Google Scholar * Zeng, Y. & Cullen, B. R. Sequence
requirements for micro RNA processing and function in human cells. RNA 9, 112–123 (2003). Article CAS PubMed PubMed Central Google Scholar * Farazi, T. A., Spitzer, J. I., Morozov, P.
& Tuschl, T. X. miRNA in human cancer. J. Pathol. 223, 102–15 (2011). Article CAS PubMed Google Scholar * Guo, H., Ingolia, N. T., Weissman, J. S. & Bartel, D. P. Mammalian
microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). Article ADS CAS PubMed PubMed Central Google Scholar * Lee, Y. S. & Dutta, A. MicroRNAs:
small but potent oncogenes or tumor suppressors. Curr. Opin. Investig. Drugs 7, 560–564 (2006). CAS PubMed Google Scholar * Lai, E. C. Micro RNAs are complementary to 3′ UTR sequence
motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30, 363–364 (2002). Article CAS PubMed Google Scholar * Saetrom, O., Snøve, O., Jr & Saetrom, P. Weighted
sequence motifs as an improved seeding step in microRNA target prediction algorithms. RNA 11, 995–1003 (2005). Article CAS PubMed PubMed Central Google Scholar * Meltzer, P. S. Cancer
genomics: small RNAs with big impacts. Nature 435, 745–746 (2005). Article ADS CAS PubMed Google Scholar * Calin, G. A. et al. MicroRNA profiling reveals distinct signatures in B cell
chronic lymphocytic leukemias. Proc Natl Acad Sci USA 101, 11755–11760 (2004). Article ADS CAS PubMed PubMed Central Google Scholar * Lu, J. et al. MicroRNA expression profiles
classify human cancers. Nature 435, 834–838 (2005). Article ADS CAS PubMed Google Scholar * Li, Z., Yang, C. S., Nakashima, K. & Rana, T. M. Small RNA-mediated regulation of iPS
cell generation. EMBO J. 30, 823–834 (2011). Article PubMed PubMed Central CAS Google Scholar * Lin, S. L. et al. Mir-302 reprograms human skin cancer cells into a pluripotent
ES-cell-like state. RNA 14, 2115–2124 (2008). Article CAS PubMed PubMed Central Google Scholar * Lowery, A. J. et al. MicroRNA signatures predict oestrogen receptor, progesterone
receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 11, R27 (2009). Article PubMed PubMed Central CAS Google Scholar * Miura, N. et al. A noncoding RNA gene on
chromosome 10p15.3 may function upstream of hTERT. BMC Mol. Biol. 2, 10:5 (2009). Google Scholar * Panopoulos, A. D. et al. The metabolome of induced pluripotent stem cells reveals
metabolic changes occurring in somatic cell reprogramming. Cell Res. 22, 168–177 (2012). Article CAS PubMed Google Scholar * Ghildiyal, M. & Zamore, P. D. Small silencing RNAs: an
expanding universe. Nat. Rev. Genet. 10, 94–108 (2009). Article CAS PubMed PubMed Central Google Scholar * Ladeiro, Y. et al. MicroRNA profiling in hepatocellular tumors is associated
with clinical features and oncogene/tumor suppressor gene mutations. Hepatology 47, 1955–1963 (2008). Article CAS PubMed Google Scholar * Farazi, T. A., Hoell, J. I., Morozov, P. &
Tuschl, T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 774, 1–20 (2013). Article CAS PubMed PubMed Central Google Scholar * He, L. et al. A microRNA polycistron as a potential human
oncogene. Nature 435, 828–833 (2005). Article ADS CAS PubMed PubMed Central Google Scholar * Goodarzi, H., Elemento, O. & Tavazoie, S. Revealing global regulatory perturbations
across human cancers. Mol. Cell 36, 900–911 (2009). Article CAS PubMed PubMed Central Google Scholar * Okita, K., Ichisaka, T. & Yamanaka, S. Generation of germline-competent
induced pluripotent stem cells. Nature 448, 313–317 (2007). Article ADS CAS PubMed Google Scholar * Fujioka, T. et al. Establishment of induced pluripotent stem cells from human
neonatal tissues. Hum. Cell 23, 113–118 (2010). Article PubMed Google Scholar * Yehezkel, S. et al. Reprogramming of telomeric regions during the generation of human induced pluripotent
stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics 21, 6 (2010). Google Scholar * Bhutani, N. et al. Reprogramming towards pluripotency requires
AID-dependent DNA demethylation. Nature 463, 1042–1047 (2010). Article ADS CAS PubMed PubMed Central Google Scholar * De Carvalho, D. D., You, J. S. & Jones, P. A. DNA methylation
and cellular reprogramming. Trends Cell Biol. 20, 609–617 (2010). Article CAS PubMed PubMed Central Google Scholar * Saito, Y. et al. Increased protein expression of DNA
methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int. J. Cancer 105, 527–532 (2003). Article CAS
PubMed Google Scholar * Tatemichi, M., Hata, H. & Nakadate, T. Ectopic expression of activation-induced cytidine deaminase caused by epigenetics modification. Oncol. Rep. 25, 153–158
(2011). CAS PubMed Google Scholar * Gunaratne, P. H. Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr. Stem Cell Res. Ther.
4, 168–177 (2009). Article CAS PubMed Google Scholar * Sen, G. L. et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463, 563–567 (2010). Article ADS
CAS PubMed PubMed Central Google Scholar * Nascimento, E. M., Cox, C. L., MacArthur, S., Hussain, S., Trotter, M. et al. The opposing transcriptional functions of Sin3a and c-Myc are
required to maintain tissue homeostasis. Nat. Cell Biol. 13, 1395–1405 (2011). Article CAS PubMed PubMed Central Google Scholar * Lin, S. L. et al. Regulation of somatic cell
reprogramming through inducible mir-302 expression. Nucleic Acids Res. 39, 1054–1065 (2010). Article ADS PubMed PubMed Central CAS Google Scholar * Kawamura, T. et al. Linking the p53
tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 (2009). Article ADS CAS PubMed PubMed Central Google Scholar * Zhu, Z. et al. Cancer stem/progenitor
cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int. J. Cancer 126, 2067–2078 (2010). Article CAS PubMed Google Scholar * Henry, J. C. et al. miR-199a-3p
targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 403, 120–125 (2010). Article CAS PubMed PubMed Central Google
Scholar * Liu, T. et al. High efficiency of reprogramming CD34+ cells derived from human amniotic fluid into induced pluripotent stem cells with Oct4. Stem Cells Dev. 21, 2322–2332 (2012).
Article CAS PubMed Google Scholar * Sand, M. et al. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest. 28, 649–653 (2010).
Article CAS PubMed Google Scholar * Chen, M. et al. Serum starvation induced cell cycle synchronization facilitates human somatic cells reprogramming. PLoS One 7, e28203 (2012). Article
ADS CAS PubMed PubMed Central Google Scholar * Plaisant, M. et al. Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells 27,
703–713 (2009). Article CAS PubMed Google Scholar * Tsai, C. C. et al. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem
cells. Molecular Cell 47, 169–182 (2012). Article CAS PubMed Google Scholar * Takai, A. et al. A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53
mutations. Oncogene 28, 1469–1478 (2009). Article CAS Google Scholar * Han, J., Sachdev, P. S. & Sidhu, K. S. A combined epigenetic and non-genetic approach for reprogramming human
somatic cells. PLoS One 5, e12297 (2010). Article ADS PubMed PubMed Central CAS Google Scholar * Mitchell, N. E. et al. Real-time methylomic aberrations during initiation and
progression of induced human mammary epithelial cell tumorigenesis. Epigenomics 5, 155–165 (2013). Article CAS PubMed Google Scholar * Zhang, D. et al. AMD1 is essential for ESC
self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 26, 461–473 (2012). Article CAS PubMed PubMed Central Google Scholar *
Hirayama, A. et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 69, 4918–4925
(2009). Article CAS PubMed Google Scholar * Nakagawa, M. et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106
(2008). Article CAS PubMed Google Scholar * Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). Article
CAS PubMed Google Scholar * Miura, N. et al. Clinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor
marker for lung cancer. Cancer Sci. 97, 1366–1373 (2006). Article CAS PubMed Google Scholar * Shomori, K. et al. Thymidine phosphorylase expression in human colorectal mucosa, adenoma
and carcinoma: role of p53 expression. Pathol. Int. 49, 491–499 (1999). Article CAS PubMed Google Scholar * Ohashi, Y. et al. Depiction of metabolome changes in histidine-starved
Escherichia coli by CE-TOFMS. Mol. Biosyst. 4, 135–147 (2008). Article CAS PubMed Google Scholar * Baran, R. et al. MathDAMP: a package for differential analysis of metabolite profiles.
BMC Bioinformatics 13, 7:530 (2006). Google Scholar * Klukas, C. & Schreiber, F. Integration of -omics data and networks for biomedical research with VANTED. J. Integr. Bioinform. 2,
7:112 (2010). Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by a Grant-in-Aid of Research for Promoting Technological Seeds B (development type), the Takeda
Science Foundation, the Princess Takamatsu Cancer Research Fund, the Adaptable and Seamless Technology Transfer Program through Target-driven R&D (Exploratory Research) of the Japan
Science and Technology Agency (JST) and a JSPS KAKENHI Grant (Grant-in-Aid for Challenging Exploratory Research) Number 23659285; none of these sources present a conflict of interest. The
cell lines used in this study were provided by the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University, Japan and the American Type
Culture Collection (ATCC). All PCR primers were designed by the INTEC Web and Genome Informatics Corporation (Tokyo, Japan). We thank Dr. Satoshi Kuwamoto regarding the pathological
examination of xenografted tumor cells. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Division of Pharmacotherapeutics, Department of Pathophysiological & Therapeutic Science, 86
Nishicho, Yonago, Tottori, 683-8503, Japan Satoshi Tsuno, Xinhui Wang, Junichi Hasegawa & Norimasa Miura * Division of Organ Pathology, Tottori University, 86 Nishicho, Yonago, Tottori,
683-8503, Japan Kohei Shomori Authors * Satoshi Tsuno View author publications You can also search for this author inPubMed Google Scholar * Xinhui Wang View author publications You can also
search for this author inPubMed Google Scholar * Kohei Shomori View author publications You can also search for this author inPubMed Google Scholar * Junichi Hasegawa View author
publications You can also search for this author inPubMed Google Scholar * Norimasa Miura View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
The author(s) have made the following declarations regarding their contributions: N.M. conceived and designed the experiments. S.T. and X.W. performed the experiments. S.T., K.S. and N.M.
analyzed the data. S.T. and X.W. contributed reagents/materials/analytical tools. S.T., J.H. and N.M. wrote the manuscript. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing financial interests. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY INFORMATION Supplementary Figure SUPPLEMENTARY INFORMATION FS 7 video RIGHTS AND PERMISSIONS This work is
licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/ Reprints
and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Tsuno, S., Wang, X., Shomori, K. _et al._ Hsa-miR-520d induces hepatoma cells to form normal liver tissues via a stemness-mediated
process. _Sci Rep_ 4, 3852 (2014). https://doi.org/10.1038/srep03852 Download citation * Received: 05 August 2013 * Accepted: 06 January 2014 * Published: 24 January 2014 * DOI:
https://doi.org/10.1038/srep03852 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Will My Part-Time Job Affect My Medicare Premiums?1:01 Videos de AARP Will My Part-Time Job Affect My Medicare Premiums? Facebook Twitter LinkedIn Making more money affec...
Robinson r22 beta, g-hrbs, 28 september 2010ROBINSON R22 BETA, G-HRBS, 28 SEPTEMBER 2010 CONTENTS * Robinson R22 Beta, G-HRBS * Summary * Download report ROBINSON R...
Associations of plasma phospholipid cis-vaccenic acid with insulin resistance markers in non-diabetic men with hyperlipidemiaABSTRACT BACKGROUND The role of fatty acids (FA) in the pathogenesis of insulin resistance and hyperlipidemia is a subje...
The wash-up from the canning byelectionIn Saturday’s byelection in the federal electorate of Canning, the Liberal Party retained the seat with a swing of 6-7% ...
What are the various kinds of french residency cards?PEOPLE FROM THE US AND OTHER COUNTRIES OUTSIDE THE SCHENGEN AREA - INCLUDING BRITONS POST-BREXIT - CAN LIVE, WORK AND ST...
Latests News
Hsa-mir-520d induces hepatoma cells to form normal liver tissues via a stemness-mediated processABSTRACT The human ncRNA gene _RGM249_ regulates the extent of differentiation of cancer cells and the conversion of 293...
Ihtm19000 - capital debts due to the estate: contents - hmrc internal manualIHTM19000 - CAPITAL DEBTS DUE TO THE ESTATE: CONTENTS CAPITAL DEBTS DUE TO THE ESTATE * IHTM19010 Introduction CAPITAL D...
Dow posts worst week since oct. 2008Stocks closed modestly higher in a choppy session Friday as investors snapped up beaten-down sectors following the previ...
Baltimore port shutdown stokes neighborhood pollution fearsWhile President Joe Biden’s visit to Baltimore last week brought national attention to the wreckage of the Francis Scott...
Restacking the deck: gender discrepancies in academic stem fieldsIn the fields of science, technology, engineering and math, often referred to as STEM fields, men can outnumber women 5-...