The cbs/h2s signalling pathway regulated by the carbon repressor crea promotes cellulose utilization in ganoderma lucidum
The cbs/h2s signalling pathway regulated by the carbon repressor crea promotes cellulose utilization in ganoderma lucidum"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Cellulose is an important abundant renewable resource on Earth, and the microbial cellulose utilization mechanism has attracted extensive attention. Recently, some signalling
molecules have been found to regulate cellulose utilization and the discovery of underlying signals has recently attracted extensive attention. In this paper, we found that the hydrogen
sulfide (H2S) concentration under cellulose culture condition increased to approximately 2.3-fold compared with that under glucose culture condition in _Ganoderma lucidum_. Further evidence
shown that cellulase activities of _G. lucidum_ were improved by 18.2-27.6% through increasing H2S concentration. Then, we observed that the carbon repressor CreA inhibited H2S biosynthesis
in _G. lucidum_ by binding to the promoter of _cbs_, a key gene for H2S biosynthesis, at “CTGGGG”. In our study, we reported for the first time that H2S increased the cellulose utilization
in _G. lucidum_, and analyzed the mechanism of H2S biosynthesis induced by cellulose. This study not only enriches the understanding of the microbial cellulose utilization mechanism but also
provides a reference for the analysis of the physiological function of H2S signals. SIMILAR CONTENT BEING VIEWED BY OTHERS HOW CARBON SOURCES DRIVE CELLULOSE SYNTHESIS IN TWO
_KOMAGATAEIBACTER XYLINUS_ STRAINS Article Open access 03 September 2024 EARLY CELLULAR EVENTS AND POTENTIAL REGULATORS OF CELLULASE INDUCTION IN _PENICILLIUM JANTHINELLUM_ NCIM 1366 Article
Open access 28 March 2023 A METAGENOMIC ‘DARK MATTER’ ENZYME CATALYSES OXIDATIVE CELLULOSE CONVERSION Article Open access 12 February 2025 INTRODUCTION Cellulose acts as the major component
of plant cell walls as well as an abundant renewable carbohydrate1,2. Because resource is currently being exhausted, further effective utilization of cellulose is of great significance.
Cellulose can be utilized by microorganisms as a low-cost and sustainable source of carbon1,3. Thus, microbial cellulose utilization is beneficial to the sustainable use of energy and carbon
cycles in the biosphere, and deserves emphasis. The present work provided a fundamental understanding of microbial cellulose utilization components, including cellulases, regulating
transcription factors and regulating signaling pathways. Enzymes with cellulose hydrolysis functions include three types, cellobiohydrolases (CBHs), endoglucanases (EGs) and β-glucosidases
(Bgs)4,5. Some transcription factors have also been reported to play roles in cellulose utilization in filamentous fungi, such as the positive transcriptional activators, XlnR, ClrA and
ClrB6,7,8,9, and the repressor, CreA3,10,11,12,13. Cellulase activity can be regulated by basal levels of enzyme production and transcription factors, which are widely recognized. However,
the role of many underlying regulators involved in cellulose utilization remains to be explored. To date, some signal transduction pathways have been reported to participate in the
regulation of cellulase activities under different stimuli conditions with a variety of modes, such as the G protein signaling pathway, AMP-activated protein kinase (AMPK) signaling pathway,
mitogen-activated protein kinase (MAPK) signaling pathway, Ca2+ signaling pathway and the cyclic AMP-dependent protein kinase A (cAMP-PKA) signaling pathway11,14,15,16,17,18,19. For
example, members of MAPK signaling pathway were involved in cellulose utilization in _Trichoderma reesei_, such as the negatively regulatory, Tmk1 and Tmk2, and the positively regulatory,
Tmk315,20. The Ca2+ signaling pathway was activated by cAMP to improve cellulase expression in _T. reesei_14,21. In _Aspergillus nidulans_, PKA indirectly phosphorylated at S319 of CreA
under glucose culture condition, which resulted in inhibiting entry of CreA into the nucleus and reducing the transcriptional inhibition of cellulase19. Our previous study investigated the
involvement of some signaling pathways in cellulose utilization in _G. lucidum_. For example, GlSwi6B, a member of MAPK signal transduction pathways, significantly increased the
concentration of cytosolic Ca2+, thereby promoting the activities of cellulase and xylanase in _G. lucidum_17. Glsnf1, a member of AMP-activated protein kinase, improved cellulase activity
by reducing the transcription level of the _creA_ gene11. These works add new insights into our understanding of cellulose degradation and are of great significance for the effective
utilization of cellulose as a renewable carbohydrate resource. Therefore, exploring novel signaling molecules that regulate cellulose utilization is important. Hydrogen sulfide (H2S) is now
recognized as an endogenous signaling gasotransmitter in various species22,23. Cystathionine β-synthase (CBS), which catalyzes the condensation of homocysteine and cysteine to produce H2S24.
H2S biosynthesis can be induced by multiple stresses. In _G. lucidum_, CBS-synthesized H2S was induced by heat stress and inhibited the heat-induced secondary metabolism accumulation22. In
wine-producing _Saccharomyces cerevisiae_, H2S biosynthesis was induced in the absence of assimilable nitrogen25, which suggests that H2S could be induced by nutrient deficiency.
Furthermore, H2S exhibited multiple physiological functions in response to various stresses. For example, H2S acted as an antioxidant and antiapoptotic signal in animals26. H2S also acted as
an antioxidant signal molecule to improve the growth rate of plants under Cu2+ and heat conditions27,28. Interestingly, cysteine supplementation reduced the furfural-induced accumulation of
reactive oxygen species (ROS) and increased biomass during lignocellulosic utilization through increasing H2S concentration in _Zymomonas mobilis_29. These results implied a potential role
of H2S in cellulose utilization, while direct evidence remains unavailable. Therefore, the mechanism of H2S biosynthesis and physiological function under cellulose culture conditions need to
be explored. Fungi are well-known organic-decomposing agents, especially mushrooms, which can use cellulose substrates1. _G. lucidum_ is an important large basidiomycete with both medicinal
value and economic value. Genome sequencing studies have shown that _G. lucidum_ contains one of the largest sets of wood-breaking enzymes among basidiomycetes30. Therefore, _G. lucidum_ is
a good material for studying the regulatory mechanism of cellulose utilization. However, the role of H2S signaling pathways in cellulose utilization remains unclear. In this study, we found
that the H2S concentration was increased under cellulose culture conditions. Increasing H2S concentration through pharmacological and genetic means enhanced cellulase activity. Furthermore,
carbon repressor CreA inhibited the expression of _cbs_, a gene encoding the H2S synthetic enzyme, and the biosynthesis of H2S under cellulose culture conditions. Further research found
that CreA binds to the _cbs_ promoter at “CTGGGG”. Our study explored a novel signaling molecule, H2S, which promotes the cellulose utilization in _G. lucidum_, and analyzed the mechanism of
cellulose-induced H2S biosynthesis. It was beneficial not only to the cultivation of _G. lucidum_ but also to the utilization of the most abundant carbon resources in the biosphere. RESULTS
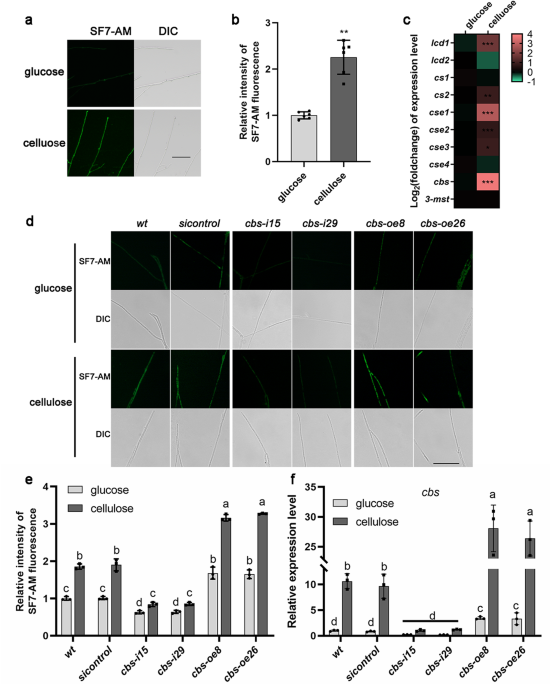
THE INTRACELLULAR CONCENTRATION OF H2S WAS INCREASED BY CELLULOSE To explore whether H2S signals respond to changes in the carbon source, H2S concentration was measured in _Ganoderma
lucidum_ under different carbon sources (glucose or microcrystalline cellulose) culture conditions. As shown in Fig. 1a, b, we observed that the fluorescence of H2S, measured by SF7-AM
fluorescence probe, was significantly (_p_ < 0.01) increased to ~2.3-folds under cellulose culture condition compared with that under glucose culture condition. This result shows that the
intracellular H2S concentration of _G. lucidum_ is increased by cellulose culture condition. The gene transcription levels of 10 putative H2S biosynthetic enzymes were also measured. As
shown in the heatmap, the expression levels of _lcd1_, _cse1_ and _cbs_ were significantly (_p_ < 0.001) increased (Fig. 1c). Among them, the _cbs_ expression level was significantly (_p_
< 0.001) increased to ~13.3-fold under cellulose culture condition compared with that under glucose culture condition (Fig. 1c), which exhibits the most pronounced response to cellulose
and implies that the cystathionine β-synthase (CBS) may be involved in regulating H2S biosynthesis under cellulose culture condition. Then, a _cbs_ gene overexpression vector was constructed
(Supplementary Fig. 1a) and transfected into _G. lucidum_. Two _cbs_-overexpressed strains (_cbs-oe8_ and _cbs-oe26_) were selected because the relative _cbs_ mRNA content increased
~3.8–4.4-fold (Supplementary Fig. 1b). The H2S concentration and the _cbs_ expression level were detected in wild-type (_wt_), _cbs_-silenced, _cbs_-overexpressed and _sicontrol_ strains
under glucose or cellulose culture conditions. The fluorescence of H2S and the expression level of _cbs_ gene in _cbs_-silenced strains significantly (_p_ < 0.05) decreased by ~54.1–54.7%
and 88.6–90.3% compared with that in the _wt_ strain under cellulose culture condition, and significantly (_p_ < 0.05) increased in _cbs_-overexpressed strains by ~70.5–76.3% and
148.7–164.6% (Fig. 1d, e, f), which indicated that CBS promoted H2S biosynthesis in _G. lucidum_ under cellulose culture condition. In addition, cellulose culture condition result in a
significant (_p_ < 0.05) increase in the fluorescence of H2S in _cbs_-silenced strains compared with glucose culture conditions to ~1.3-folds, which was less than that in the _wt_ strain,
and no significant (_p_ > 0.05) change in the expression level of _cbs_ gene (Fig. 1d, e, f). These results suggest that cellulose promotes the H2S biosynthesis in _G. lucidum_, and CBS
might be one of the main H2S biosynthetic enzymes under cellulose culture condition. H2S ENHANCED CELLULASE ACTIVITY AND CELLULOSE UTILIZATION IN _G. LUCIDUM_ To explore the effect of H2S
signal on cellulose utilization, cellulase activity was measured in the presence of sodium hydrosulfide (NaHS, a H2S donor) and hypotaurine (HT, a H2S scavenger), at a concentration that can
significantly alter the intracellular H2S content in _G. lucidum_ (Supplementary Fig. 2). As shown in Fig. 2a, the addition of NaHS increased cellulase activity, and the promoting effect
increased gradually with increasing NaHS concentration. The addition of 60 μM NaHS significantly (_p_ < 0.001) increased cellulase activity by ~27.6% compared with no treatment (Fig. 2a).
The addition of HT inhibited cellulase activity, and the inhibitory effect was more evident with increasing HT concentration (Fig. 2b). HT (2 mM) significantly (_p_ < 0.001) reduced
cellulase activity by ~18.6% compared with no treatment (Fig. 2b). Pharmacological experiments suggest that H2S enhances cellulase activity in _G. lucidum_ under cellulose culture condition.
To explore the potential influence of H2S biosynthesized by CBS on cellulose utilization, cellulase activity was measured in _wt_, _cbs_-silenced, _cbs_-overexpressed and _sicontrol_
strains in the presence of NaHS and HT. Cellulase activity in _cbs_-silenced strains significantly (_p_ < 0.001) reduced by ~31.9–32.2% compared with that in the _wt_ strain (Fig. 2c).
NaHS addition significantly (_p_ < 0.01) increased cellulase activity of _cbs_-silenced strains by ~27.1–28.2% compared with under cellulose culture condition alone (Fig. 2c). Cellulase
activity in _cbs_-overexpressed strains significantly (_p_ < 0.001) increased by ~18.2–18.7% compared with that in the _wt_ strain (Fig. 2d). HT addition significantly (_p_ < 0.001)
reduced cellulase activity in _cbs_-overexpressed strains by ~17.8-18.6% with under only cellulose culture condition (Fig. 2d). To demonstrate the potential influence of CBS on cellulose
utilization of _G. lucidum_, wood chips were used as a main carbon source to cultivate _wt_, _cbs_-silenced, _cbs_-overexpressed and _sicontrol_ strains for 15 days, and the growth length
were measured. As shown in Fig. 2e, f, the growth length of _cbs_-silenced strains significantly (_p_ < 0.001) decreased by ~53.4–56.1% compared with _wt_ strain, while
_cbs_-overexpressed significantly (_p_ < 0.01) increased by ~14.4–16.4%. These results suggest that CBS-synthesized H2S improves the cellulase activity and cellulose utilization. These
combined results suggest that H2S improves the cellulase activity of _G. lucidum_ under cellulose culture condition. Y1H SCREENING FOR REGULATORS OF THE _CBS_ GENE IDENTIFIES DIVERSE
TRANSCRIPTION FACTORS To further understand the mechanism of cellulose induced H2S biosynthesis, _cbs_ promoter was analyzed. The intervening region between the _cbs_ gene and upstream gene
is very short (551 bp). Therefore, a total potential _cbs_ promoter (+1 to −551 pb) was used as bait in a yeast one-hybrid (Y1H) library screen. The Y1H assay indicated that a total of 13
putative transcription factors (TFs) may directly bind to the _cbs_ promoter (Supplementary Table 1). These putative TFs might regulate _cbs_ gene transcription and H2S biosynthesis in _G.
lucidum_ under different conditions. Among them, three TFs, had been reported in _G. lucidum_, were observed: CreA (mediated cellulose utilization of _G. lucidum_11), GCN4 and SKO1 (mediated
nitrogen utilization of _G. lucidum_31), and four highly conserved TFs were observed: TFIIB, MCM1, Xbp1, and Crz1 (Fig. 3a and Supplementary Table 1). To explore the regulatory mechanism of
_cbs_ transcription and H2S biosynthesis under cellulose culture condition in _G. lucidum_, CreA, a classical carbon catabolite repressor, was selected for further study. CREA COULD BIND TO
THE _CBS_ PROMOTER To further determine the binding effect of CreA to the _cbs_ promoter, Y1H assay and EMSA were performed. The Y1H assay indicated that compared with the negative control
(pGADT7), Y1HGold yeast transformed pGADT7-CreA, developed obvious colonies (Fig. 3c). The EMSA result showed that compared with lanes without protein addition, an obvious binding complex
band was observed in lanes with purified His-CreA protein addition (Fig. 3d). These binding effects were competitively inhibited by unlabeled cold probes (Fig. 3d). Gradually decreased
binding complex bands were observed between 5’ biotin-labeled probes of the _cbs_ promoter and purified His-CreA protein in lanes with a decrease in protein concentration (100 mM, 50 mM, 20
mM) (Fig. 3d). These results suggest that CreA can bind to the _cbs_ promoter. To further explore the binding site of CreA in the _cbs_ promoter, the _cbs_ promoter was analyzed, and
observed a typical CreA binding site, “CTGGGG”, at -232 bp to -226 bp (Fig. 3b). Then, two GG nucleotides in the middle of the conserved binding domain were mutated to AA nucleotides as a
previous study32. As shown in results of the Y1H assay, compared with the positive control (CBS-AbAi), Y1HGold yeast, transformed CBS-Mut-AbAi, did not develop significant colonies (Fig.
3c). The EMSA showed that compared with the positive control (non-mutated 5’ biotin-labeled probe), the mutated 5’ biotin-labeled probe could not bind with purified CreA protein (Fig. 3d).
These results indicate that CreA can bind to the _cbs_ promoter at “CTGGGG”. CREA INHIBITS _CBS_ GENE EXPRESSION AND REDUCES H2S BIOSYNTHESIS To explore the regulatory role of CreA in the
CBS/H2S signaling pathway in _G. lucidum_, a _CreA_-overexpress vector was constructed (Supplementary Fig. 3a) and transfected into _G. lucidum_. Two _creA_-overexpressed strains (_creA-oe6_
and _creA-oe8_) were selected for further analyses. The relative mRNA content level significantly (_p_ < 0.001) increased about 5.7–6.0-fold and the relative protein content level
significantly (_p_ < 0.001) increased by ~52.7–54.5% in _creA_-overexpressed strains compared with the _wt_ strain (Supplementary Fig. 3b, Supplementary Fig. 6, Fig. 4a, b). The relative
mRNA and protein content levels in _creA_-silenced strains, previously established22, significantly (_p_ < 0.001) reduced by ~74.8–75.6% and 32.6–33.7% compared with those in the _wt_
strain, respectively (Supplementary Fig. 3b, Supplementary Fig. 6, Fig. 4a, b). Transcription levels of the _cbs_ gene and fluorescence levels of H2S were measured in _wt_, _creA_-silenced,
_creA_-overexpressed and _sicontrol_ strains. Compared with that in the _wt_ strain, the transcription level of the _cbs_ gene in _creA_-silenced strains significantly (_p_ < 0.001)
increased to ~2.1-folds and the fluorescence of H2S was significantly (_p_ < 0.01) increased by ~47.5–49.2% (Fig. 4c, d, e). The transcription level of the _cbs_ gene and the fluorescence
of H2S in _creA_-overexpressed strains significantly (_p_ < 0.01) decreased by ~44.3–48.3% and 29.1–30.3%, respectively (Fig. 4c, d, e). These results indicate that CreA reduces _cbs_
gene transcription levels and intracellular H2S biosynthesis. CREA INHIBITS _CBS_ GENE EXPRESSION AND REDUCES H2S BIOSYNTHESIS UNDER CELLULOSE CULTURE CONDITION To explore the role of CreA
in _cbs_ expression and H2S biosynthesis, the binding effect of CreA to the _cbs_ promoter was measured under glucose and microcrystalline cellulose culture condition. From the results of
ChIP-qPCR, we found that cellulose culture condition significantly (_p_ < 0.05) reduced the binding of CreA to the _cbs_ promoter in the _wt_ strain by ~51.1% compared with glucose
culture condition (Supplementary Fig. 4 and Fig. 5a), indicating that cellulose culture conditions inhibited the DNA binding activity to the _cbs_ promoter of CreA. In addition, under
cellulose culture condition, the binding of CreA to the _cbs_ promoter was significantly (_p_ < 0.05) reduced by ~56.9–64.4% in _creA_-silenced strains and significantly (_p_ < 0.05)
increased to ~3.9–4.2-fold in _creA_-overexpressed strains compared with that in the _wt_ strain (Fig. 5a). Then, transcription levels of the _cbs_ gene and fluorescence levels of H2S were
measured in the _wt_, _creA_-silenced, _creA_-overexpressed and _sicontrol_ strains. Under cellulose culture condition, the transcription level of the _cbs_ gene in _creA_-silenced strains
was significantly (_p_ < 0.05) increased to ~2.0-folds compared with that in the _wt_ strain, and the fluorescence of H2S was significantly (_p_ < 0.05) increased to ~1.8-folds (Fig.
5b, c, d). The transcription level of the _cbs_ gene in _creA_-overexpressed strains under cellulose culture condition was significantly (_p_ < 0.05) reduced by ~85.9–86.5% compared with
that in the _wt_ strain under, and the fluorescence of H2S was significantly (_p_ < 0.05) reduced by ~68.8–70.8% (Fig. 5b, c, d). These results indicate that cellulose inhibit the DNA
binding activity to the _cbs_ promoter of CreA, thereby enhancing _cbs_ transcription and H2S biosynthesis under cellulose culture conditions. CREA INHIBITS THE POSITIVE REGULATION OF
CBS/H2S IN CELLULOSE UTILIZATION OF _G. LUCIDUM_ To explore the role of H2S and CreA in cellulose utilization of _G. lucidum_, a _creA_-_cbs_-silenced plasmid was constructed (Supplementary
Fig. 5a). Two _creA_-_cbs_-silenced strains (_creA-cbs-i23_ and _creA-cbs-i27_) were selected because of ~53.4–55.7% and 53.3–58.2% significant (_p_ < 0.01) reduction in the relative mRNA
content levels of the _creA_ and _cbs_ genes, respectively (Supplementary Fig. 5b, c). The transcription level of the _cbs_ gene and the fluorescence of H2S in _creA-cbs-i23_ and
_creA-cbs-i27_ strains under cellulose culture condition significantly (_p_ < 0.01) reduced by ~85.6–86.0% and 74.1–77.5% compared with those in _creA_-silenced strains, respectively, but
no significant (_p_ > 0.05) difference was observed compared with those in _cbs_-silenced strains (Fig. 5b, c, d). Cellulase activity in the _wt_, _creA_-silenced, _creA_-overexpressed,
_creA_-_cbs_-silenced and _sicontrol_ strains was measured in the presence of NaHS and HT treatment. Cellulase activity in _creA_-silenced strains was significantly (_p_ < 0.05) increased
by ~54.6–58.1% compared with that in the _wt_ strain, and significantly (_p_ < 0.05) decreased by ~35.6–38.2% in _creA_-overexpressed strains (Fig. 6a). This result suggested that CreA
inhibits cellulase activity. When _creA_-overexpressed strains treated with NaHS, the cellulase activity significantly (_p_ < 0.05) increased by ~27.6–29.6% compared with that no
treatment (Fig. 6a). Cellulase activity in _creA_-_cbs_-silenced strains were significantly (_p_ < 0.001) reduced by ~59.7–61.7% compared with that in _creA_-silenced strains, but there
was no significant difference compared with that in _cbs_-silenced strains (Fig. 6a). These results suggest that the silencing of both _creA_ and _cbs_ can inhibit the _creA_
silencing-induced decrease in cellulase activity. Then, the growth rates of the _wt_, _creA_-silenced, _creA_-overexpressed, _creA_-_cbs_-silenced and _sicontrol_ strains on wood chips were
further tested. As shown in the Fig. 6b, c, the growth rates of the _creA_-overexpressed and _creA_-_cbs_-silenced strains were significantly (_p_ < 0.05) reduced by ~52.0–54.4% and
51.4–53.1% compared to that of the _wt_ strain, while the growth rate of the _creA_-silenced strain was significantly (_p_ < 0.05) increased by ~22.1–23.1%. These results suggest that
CreA reduces cellulose utilization of _G. lucidum_ by inhibiting CBS/H2S signaling pathway. DISCUSSION Cellulose utilization contributes to the carbon cycle in the biosphere. Microbial
cellulose utilization is widely used in both agriculture and industry17,33. Exploring the potential molecular regulatory mechanisms of cellulose utilization in _Ganoderma lucidum_, a
medicinal and edible basidiomycete, is not only be crucial to understanding the mechanism of mushroom growth and development, but also provides a theoretical basis for other
cellulose-utilizing microorganisms. The efficiency of cellulose utilization dependent on the regulation of transcription factors and the activity of cellulase is widely recognized. However,
recent studies have found that some signaling pathways could also take part in the regulation of cellulose utilization, such as Ca2+, cAMP and MAPK signals14,15,17. Therefore, the underlying
molecular regulatory mechanism of cellulose utilization in _G. lucidum_ deserves emphasis and awaits further investigation. In this paper, we depict the promoting effect of hydrogen sulfide
(H2S) on the activity of cellulase in _G. lucidum_ for the first time. Furthermore, H2S biosynthesis was negatively regulated by the carbon repressor transcription factor, CreA. Our results
provided new insights into the mechanism of microbial cellulose utilization. In this work, we observed that H2S improved the cellulose utilization in _G. lucidum_. Before this work, the
role of other signaling molecules on cellulase activity and its mechanism were also reported, such as cAMP and Ca2+14,17,21,34. For instance, cAMP and Ca2+ improved the expression of
cellulase in _Trichoderma reesei_14,21. Increasing cytosolic Ca2+ concentration promoted the activities of cellulase and xylanase in _G. lucidum_17. The silence of _PoLaeA2_ and _PoLaeA3_,
key global regulators, leads to a reduction in cytosolic Ca2+ content and cellulase activity in _Pleurotus ostreatus_34. In addition, H2S has also been widely reported to have complex
interactions with various signal molecules. For example, H2S increases cAMP level in resting cells, while decreases cAMP level when adenylyl cyclases are activated35. Additionally, H2S also
acts as a regulator of the Ca2+ channels to regulate the intracellular Ca2+ concentration in various animal cells with different physiological effects36,37,38. H2S enhance the tolerance of
plants to drought stress by activating Ca2+ signaling39. Our previous work demonstrated that H2S leads to a decrease in cytosolic Ca2+ level in _G. lucidum_ under heat stress22. Therefore,
whether H2S improve cellulase activity under cellulose culture conditions through interactions with other signaling pathways remains to be further studied. In our previous work, we found
that the silencing of _creA_ led to an increase in transcription levels of cellulase genes and cellulase activity in _G. lucidum_ under cellulose culture condition11. In this study, we
observed that the cellulase activity in the _creA_-_cbs_ silenced strains were significantly decreased than that of the _wt_ strain (Fig. 6a). These results imply an important regulatory
role of CBS-synthesized H2S in cellulase activity. Previous studies have reported that H2S affect protein function by a variety of potential mechanisms. Firstly, H2S could directly regulate
the activity of proteins by persulfidation. For example, H2S regulates autophagy under endoplasmic reticulum stress in _Arabidopsis_ by persulfidating ATG18a, a core autophagy component40.
Secondly, H2S could regulate transcription factors by persulfidation, thereby affect the transcription expression of multiple genes downstream of the transcription factor. For instance, H2S
involve in the regulation of cucurbitacin C synthesis in cucumber by increasing the persulfidation level of His-Csa5G156220 and His-Csa5G157230 (transcription factors) and transcriptionally
activate Csa6G088690 (a key synthetase for CuC generation)41. Thirdly, H2S also could regulate the upstream regulator of transcription factors by persulfidation, thereby indirectly affect
the downstream transcription expression of multiple genes. For example, H2S regulates ABA signaling by persulfidating SnRK2.6, promoting the activity of SnRK2.6 and its interaction with a
transcription factor acting downstream of ABA signaling, ABF2, in guard cells42. In addition, these various mechanisms by which intracellular H2S affects physiological processes often exist
simultaneously. For instance, in plants, H2S not only regulated transcription levels of drought-responsive genes but also induced the persulfidation level of proteins, which involved in
cellular response to oxidative stress, hydrogen peroxide catabolism and so on, to involve in drought stress responses43. Therefore, further research is needed to clarify regulatory
mechanisms of H2S on cellulase activity in _G. lucidum_. Intracellular H2S biosynthesis was regulated by some transcription factors and induced by stress44. For example, transcription factor
specificity protein 1 (Sp1) can bind to the promoter of both _cse_ and _cbs_ gene, two H2S biosynthetic enzymes, to regulate H2S biosynthesis in animals45,46. Nuclear factor (NF)-Y and
upstream stimulatory factor 1 (USF-1) were also involved in the regulation of _cbs_ promoter activity in HepG2 cells47. The transcription factor OsNACL35 increased in H2S concentration by
directly upregulating the expression of _OsDCD1_ by binding to the promoter of _OsDCD1_ gene under salinity stress48. The regulation of transcription levels of H2S biosynthetic enzymes had
been reported in animals and plants, but less so in microorganisms. In this study, yeast one-hybrid (Y1H) library screening results revealed the underlying transcription factors of the _cbs_
gene, including CreA (mediated cellulose utilization of _G. lucidum_11), GCN4 and SKO1 (mediated nitrogen utilization of _G. lucidum_31), and four putative transcription factors (TFIIB,
MCM1, Xbp1, Crz1). We further revealed that CreA combined with the _cbs_ promoter, transcriptionally provoked _cbs_ gene expression and improved H2S biosynthesis under cellulose culture
condition by pharmacological experiments and genetic experiments. The roles of other transcription factors need to be further studied. The potential binding of multiple transcription factors
to the _cbs_ gene implied that H2S may be induced by a variety of growing conditions in _G. lucidum_, and providing unlimited possibilities for the physiological function of H2S signals
under stress. CreA, a carbon catabolite repressor, was previously reported to directly bind to the gene promoter of cellulolytic enzymes, inhibit the transcription of related genes, and
regulate cellulose utilization in filamentous fungi, such as _T. reesei_, _Aspergillus nidulans_, _G. lucidum_ and _Neurospora crassa_3,11,12,49. The transcriptomics analysis and the
secretome analysis in _N. crassa_ were observed that CreA repressed the expression level of genes, encoding enzymes involved in the utilization of alternative carbon sources, and cellulase
activity under cellulose culture condition50. In _T. reesei_, CreA was considered as a regulator of the glucose assimilation rate51. Recent research has also shown that CreA could bind not
only to the promoter of cellulose related genes, but also to the promoter of other genes to play a primary role in diverse physiological processes, such as carbon metabolism, secondary
metabolism, iron homeostasis, oxidative stress response, development, N-glycan production, unfolded protein response, and nutrient and ion transport in _A. nidulans_52. Beyond carbon
metabolism regulation, studies in various fungal species indicate additional CreA functions. For example, the ΔcreA mutants of _Aspergillus flavus_53, _Magnaporthe oryzae_54, and _Beauveria
bassiana_55 are affected in development and virulence, while repression of genes with functions in nitrogen uptake, development, chromatin remodeling, and the mediator complex depends on a
functional CreA in _T. reesei_51. In _Aspergillus fumigatus_, the ΔcreA mutant impacts growth, fitness, and virulence56. In this study, we found that CreA negatively regulated _cbs_
expression by binding to the _cbs_ promoter, and inhibiting H2S biosynthesis, thereby reducing cellulase activity. Additionally, under repressing carbon sources culture condition, such as
cellulose, the transcriptional level of CreA was decreased32,49. Therefore, CBS/H2S signaling pathway was provoked under cellulose culture condition in _G. lucidum_. H2S is a signaling
molecule with multiple biological functions, and these results also implied that CreA may play a greater biological role through the regulation of H2S biosynthesis. In this study, we
revealed that H2S signals were promoted by activating _cbs_ transcription level under cellulose culture condition (Fig. 7). Then, we observed that H2S improved cellulase activity in _G.
lucidum_ (Fig. 7). Furthermore, we observed that CreA could bind to the promoter of _cbs_ gene and reduce the transcriptional level of _cbs_ gene (Fig. 7). In the case of repressing carbon
sources, such as cellulose culture condition, CreA mRNA levels were decreased32,49. Therefore, cellulose reduced the inhibitory effect of CreA on the transcription level of _cbs_ gene,
thereby activating CBS/H2S signaling pathway under cellulose culture condition. These results explored a novel signaling molecule, H2S, which participate in the regulation of cellulose
utilization. Our study not only provides insight into the response mechanism of H2S signals in exposure to cellulose in _G. lucidum_ but also benefits to the utilization of the most abundant
energy resources in the biosphere. METHODS STRAINS AND CULTURE CONDITIONS The wild-type (_wt_) _Ganoderma lucidum_ strain (obtained from Shanghai Academy of Agricultural Science) was used
in a previous work22. The _G. lucidum_ strains used in this experiment: _sicontrol_, _cbs_-silenced and _cbs_-overexpressed strains were established previously22; _creA-_silenced strains
were established previously11; _creA-_overexpressed strains were established as described in Supplementary Fig. 1, _creA-cbs-_silenced strains were established as described in Supplementary
Fig. 3. The culture conditions of _G. lucidum_ was used as previous works22,57. For wood chips culture, _G. lucidum_ were cultured on edible mushroom cultivation material (60% wood chip, 20%
cottonseed hull, 18% wheat bran, 1% sucrose and 1% gypsum) at 28 °C and for 15 days. For other detection, _G. lucidum_ were cultured in glucose-containing nutrient-rich liquid CYM medium at
28°C and 150 rpm for 5 days, and then collected and changed into single carbon source (1% glucose or microcrystalline cellulose) liquid MCM medium (0.46% KH2PO4, 0.05% MgSO4-7H2O, 0.5%
(NH4)2SO4, 2 ml/l trace elements) at 28°C and 150 rpm for 2 days. TRANSCRIPTION LEVEL DETECTION The transcription level of _cbs_ and _creA_ were detected by RT-qPCR according to method
described previously22. Total RNAiso Plus (TaKaRa, Dalian, China) was used to extract total RNA and cDNA was reverse transcribed using a PrimeScript RT reagent kit (TaKaRa, Dalian, China).
18S rRNA was used as a reference to analyze the transcription levels of _cbs_ and _creA_. The oligonucleotide primers used are listed in the Supplementary Table 2. The relative transcription
levels of genes were determined using the 2−ΔΔCT method. ENDOGENOUS H2S CONCENTRATION DETECTION H2S concentration in _G. lucidum_ was detected after incubation with sulfidefluor-7
acetoxymethyl ester (SF7-AM) as described in a previous study22,58. The average fluorescence intensity values of all mycelia in each photo were quantified. Zeiss Axio Imager A1 fluorescence
microscope was used to fluorescence image for all samples under the same microscopy settings. The average fluorescence intensity of mycelium was analyzed using ZEN software. ENDOCELLULASE
ACTIVITY (CMCASE) DETECTION Endocellulase activity was detected according to previously described methods11. The culture supernatants were collected for endocellulase activity (CMCase)
assays. In brief, citric acid buffer (50 mM, pH 4.8) and 2% (W/V) sodium carboxymethyl cellulose (CMC-Na) were added and the reaction was carried out at 50 °C for 30 min, and then detected
optical density at 540 nm. YEAST ONE-HYBRID ASSAY A yeast one-hybrid (Y1H) assay was performed using the matchmaker gold yeast one-hybrid library screening system (Clontech, China). The
target sequence of _cbs_ promoter (+1 to −551pb) was cloned and inserted into the pAbAi vector. The CBS-AbAi vector was digested with BstBI enzyme (TAKARA, China) and then transformed into
yeast stains. Strains were grown on SD/-Ura media for 2 days and positive transformants were selected by PCR. Next, the minimal inhibitory concentration of aureobasidin A (AbA) was confirmed
as follows. Y1HGold (CBS-AbAi) were suspended in 0.9% NaCl (OD600: 0.002) and dotted onto SD/-Ura medium with AbA (0, 100, 150, 200, 300, 400, 500, 600, 800, and 1000 ng ml−1) for 3 days.
AbA (600 ng ml−1) completely suppressed the growth of yeast strains and was used for subsequent experiments. The Y1H screen assay was performed as previously described59. The cDNA library
(pGADT7 vector) was conducted by SMART cDNA production technology (oebiotech, China). Then, screen the cDNA library by cotransformation and transformed yeast stains are plated on
SD/-Leu/+AbA to select for colonies. For the transcriptional activation test, the full-length complementary cDNA of _creA_ was cloned and inserted into the pGADT7 vector. The constructed
plasmids (pGADT7-CreA) were then transformed into Y1HGold (CBS-AbAi) on SD/-Leu/+AbA medium for 3 days, and the positive transformants and were chosen and dotted as described above.
EXPRESSION AND PURIFICATION OF CREA CreA protein was expressed and purified as previously described11. The _creA_-28a plasmid was transformed into _E. coli_ BL21 (DE3), and protein
expression was induced by 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 28 °C for 4 h. Protein verification was performed using SDS-PAGE. ELECTROPHORETIC MOBILITY SHIFT ASSAY An
electrophoretic mobility shift assay (EMSA) was performed as previously described46,59. Both forward (5′-TGGCTGGGGC-3′) and reverse (5′-GCCCCAGCCA-3′) primers of _cbs_ promoter were biotin
labeled at the 5’ end. The mutated primers were 5′-TGGCTGTTGC-3′ (forward) and 5′-GCAACAGCCA-3′ (reverse) biotin labeled at the 5’ end. For EMSA analysis, the biotin-labeled primers were
incubated with purified CreA protein (10 μg, 5 μg, 2 μg, respectively) in 5 μl binding buffer [10 mM Tris-HCl (pH 8.0), 1 mM DTT, 0.1 mM EDTA, 50 mM KCl and 5% glycerol]. In addition, the
mixture without protein was used as a negative control. CHROMATIN IMMUNOPRECIPITATION ASSAY Chromatin immunoprecipitation assay (ChIP) was performed as previously described46. In brief,
cross-linking of DNA with proteins was performed with 1% formaldehyde. The nuclei were isolated, and sheared chromatin was prepared by sonication. The sheared chromatin was
immunoprecipitated with rabbit polyclonal anti-CreA antibodies and rabbit serum (D601019, Sangon, China, negative control). Another aliquot of sheared chromatin without incubation with
antibodies was prepared as input. As shown in Sigure S2, DNA, isolated from immunoprecipitation, was detected by PCR with primers for the _cbs_ promoter region were 5′-TTGACGCGGACGGACAT-3′
(forward) and 5′-GGGAAGATGGTGGCAGAA-3′ (reverse). The relative binding efficiency of CreA to the _cbs_ promoter was detected by RT-qPCR. WESTERN BLOTTING Western blotting was performed as
previously described11. Briefly, proteins from mycelia samples were separated in a 12% (w/v) SDS-PAGE gel and transferred to polyvinylidene difluoride membranes (Bio-Rad). The primary
antibodies used to detect specific proteins in this report were anti-CreA (1:2000, rabbit polyclonal), anti-Actin (1:2000, mouse; CMCTAG) and anti-Histone3 (1:2000, AT0005, CMCTAG). ImageJ
software was used to quantify the densities of the bands. STATISTICAL ANALYSIS The statistical analyses were performed using GraphPad Prism 8.0.2 (GraphPad Software, San Diego, CA, USA). All
experimental data shown in this article were carried out in three independent samples to ensure that trends and relationships observed in cultures were reproducible. The error bars indicate
the standard deviation from the mean of triplicates. The data were analyzed using Student’s t test or Duncan’s multiple range test. The _p_ < 0.05 was considered significant. REPORTING
SUMMARY Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY All data generated or analyzed during this
study are included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request. The source data underlying the
graphs in the figure are shown in Supplementary Data 1. Uncropped western blots are in Supplementary Fig. 6. REFERENCES * Lynd, L. R., Weimer, P. J., van Zyl, W. H. & Pretorius, I. S.
Microbial cellulose utilization: fundamentals and biotechnology. _Microbiol. Mol. Biol. Rev._ 66, 506–577 (2002). Article CAS PubMed PubMed Central Google Scholar * Lynd, L. R., Wyman,
C. E. & Gerngross, T. U. Biocommodity engineering. _Biotechnol. Prog._ 15, 777–793 (1999). Article CAS PubMed Google Scholar * Lichius, A., Seidl-Seiboth, V., Seiboth, B. &
Kubicek, C. P. Nucleo-cytoplasmic shuttling dynamics of the transcriptional regulators XYR1 and CRE1 under conditions of cellulase and xylanase gene expression in _Trichoderma reesei_. _Mol.
Microbiol._ 94, 1162–1178 (2014). Article CAS PubMed PubMed Central Google Scholar * Ilmén, M., Saloheimo, A., Onnela, M. L. & Penttilä, M. E. Regulation of cellulase gene
expression in the filamentous fungus _Trichoderma reesei_. _Appl. Environ. Microbiol._ 63, 1298–1306 (1997). Article PubMed PubMed Central Google Scholar * Wu, M. H., Kao, M. R., Li, C.
W., Yu, S. M. & Ho, T. D. A unique self-truncation of bacterial GH5 endoglucanases leads to enhanced activity and thermostability. _BMC Biol._ 20, 137 (2022). Article PubMed PubMed
Central Google Scholar * Stricker, A. R., Mach, R. L. & de Graaff, L. H. Regulation of transcription of cellulases- and hemicellulases-encoding genes in _Aspergillus niger_ and
_Hypocrea jecorina_ (_Trichoderma reesei_). _Appl. Microbiol. Biotechnol._ 78, 211–220 (2008). Article CAS PubMed Google Scholar * Xia, C. et al. Introduction of heterologous
transcription factors and their target genes into _Penicillium oxalicum_ leads to increased lignocellulolytic enzyme production. _Appl. Microbiol. Biotechnol._ 103, 2675–2687 (2019). Article
CAS PubMed Google Scholar * Craig, J. P., Coradetti, S. T., Starr, T. L. & Glass, N. L. Direct target network of the _Neurospora crassa_ plant cell wall deconstruction regulators
CLR-1, CLR-2, and XLR-1. _mBio_ 6, e01452–01415 (2015). Article CAS PubMed PubMed Central Google Scholar * Huberman, L. B., Coradetti, S. T. & Glass, N. L. Network of
nutrient-sensing pathways and a conserved kinase cascade integrate osmolarity and carbon sensing in _Neurospora crassa_. _Proc. Natl Acad. Sci. USA_ 114, E8665–e8674 (2017). Article CAS
PubMed PubMed Central Google Scholar * Li, Z. et al. Synergistic and dose-controlled regulation of cellulase gene expression in _Penicillium oxalicum_. _PLoS Genet._ 11, e1005509 (2015).
Article PubMed PubMed Central Google Scholar * Hu, Y. et al. In _Ganoderma lucidum_, Glsnf1 regulates cellulose degradation by inhibiting GlCreA during the utilization of cellulose.
_Environ. Microbiol._ 22, 107–121 (2020). Article CAS PubMed Google Scholar * Lin, H., Hildebrand, A., Kasuga, T. & Fan, Z. Engineering _Neurospora crassa_ for cellobionate
production directly from cellulose without any enzyme addition. _Enzym. Microbial Technol._ 99, 25–31 (2017). Article CAS Google Scholar * Hildebrand, A., Szewczyk, E., Lin, H., Kasuga,
T. & Fan, Z. Engineering _Neurospora crassa_ for improved cellobiose and cellobionate production. _Appl. Environ. Microbiol._ 81, 597–603 (2015). Article PubMed PubMed Central Google
Scholar * Chen, Y. et al. cAMP activates calcium signalling via phospholipase C to regulate cellulase production in the filamentous fungus _Trichoderma reesei_. _Biotechnol. Biofuels_ 14,
62 (2021). Article PubMed PubMed Central Google Scholar * Wang, M. et al. Role of _Trichoderma reesei_ mitogen-activated protein kinases (MAPKs) in cellulase formation. _Biotechnol.
Biofuels_ 10, 99 (2017). Article CAS PubMed PubMed Central Google Scholar * Tisch, D., Kubicek, C. P. & Schmoll, M. New insights into the mechanism of light modulated signaling by
heterotrimeric G-proteins: ENVOY acts on _gna1_ and _gna3_ and adjusts cAMP levels in _Trichoderma reesei_ (_Hypocrea jecorina_). _Fungal Genet. Biol._ 48, 631–640 (2011). Article CAS
PubMed PubMed Central Google Scholar * Lian, L. D. et al. GlSwi6 positively regulates cellulase and xylanase activities through intracellular Ca2+ signaling in _Ganoderma lucidum_. _J.
Fungi_ 8, https://doi.org/10.3390/jof8020187 (2022). * Liu, P. et al. Enhanced cellulase production by decreasing intercellular pH through H+-ATPase gene deletion in _Trichoderma reesei_
RUT-C30. _Biotechnol. Biofuels_ 12, 195 (2019). Article PubMed PubMed Central Google Scholar * Ribeiro, L. F. C. et al. Comprehensive analysis of _Aspergillus nidulans_ PKA phosphorylome
identifies a novel mode of CreA regulation. _mBio_ 10, https://doi.org/10.1128/mBio.02825-18 (2019). * Wang, M. et al. A mitogen-activated protein kinase Tmk3 participates in high
osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in _Trichoderma reesei_. _PloS One_ 8, e72189 (2013). Article CAS PubMed PubMed Central Google
Scholar * Chen, Y. et al. N,N-dimethylformamide induces cellulase production in the filamentous fungus _Trichoderma reesei_. _Biotechnol. Biofuels_ 12, 36 (2019). Article PubMed PubMed
Central Google Scholar * Tian, J. L. et al. Hydrogen sulfide, a novel small molecule signalling agent, participates in the regulation of ganoderic acids biosynthesis induced by heat stress
in _Ganoderma lucidum_. _Fungal Genet. Biol._ 130, 19–30 (2019). Article CAS PubMed Google Scholar * Zhao, W., Zhang, J., Lu, Y. & Wang, R. The vasorelaxant effect of H2S as a novel
endogenous gaseous K(ATP) channel opener. _EMBO J._ 20, 6008–6016 (2001). Article CAS PubMed PubMed Central Google Scholar * Landry, A. P., Roman, J. & Banerjee, R. Structural
perspectives on H2S homeostasis. _Curr. Opin. Struct. Biol._ 71, 27–35 (2021). Article CAS PubMed PubMed Central Google Scholar * Jiranek, V., Langridge, P. & Henschke, P. A.
Regulation of hydrogen sulfide liberation in wine-producing _Saccharomyces cerevisiae_ strains by assimilable nitrogen. _Appl. Environ. Microbiol._ 61, 461–467 (1995). Article CAS PubMed
PubMed Central Google Scholar * Jha, S., Calvert, J. W., Duranski, M. R., Ramachandran, A. & Lefer, D. J. Hydrogen sulfide attenuates hepatic ischemia-reperfusion injury: role of
antioxidant and antiapoptotic signaling. _Am. J. Physiol. Heart Circ. Physiol._ 295, H801–806, (2008). Article CAS PubMed PubMed Central Google Scholar * Zhang, H. et al. Hydrogen
sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. _J. Integr. Plant Biol._ 50, 1518–1529 (2008). Article CAS PubMed Google Scholar *
Christou, A., Filippou, P., Manganaris, G. A. & Fotopoulos, V. Sodium hydrosulfide induces systemic thermotolerance to strawberry plants through transcriptional regulation of heat shock
proteins and aquaporin. _BMC Plant Biol._ 14, 42 (2014). Article PubMed PubMed Central Google Scholar * Yan, X. et al. Cysteine supplementation enhanced inhibitor tolerance of _Zymomonas
mobilis_ for economic lignocellulosic bioethanol production. _Bioresour. Technol._ 349, 126878 (2022). Article CAS PubMed Google Scholar * Liu, D. et al. The genome of _Ganoderma
lucidum_ provides insights into triterpenes biosynthesis and wood degradation. _PloS One_ 7, e36146 (2012). Article CAS PubMed PubMed Central Google Scholar * Lian, L. et al. GCN4
enhances the transcriptional regulation of AreA by interacting with SKO1 to mediate nitrogen utilization in _Ganoderma lucidum_. _Appl. Environ. Microbiol._ 88, e0132222 (2022). Article
PubMed Google Scholar * Strauss, J. et al. The function of CreA, the carbon catabolite repressor of _Aspergillus nidulans_, is regulated at the transcriptional and post-transcriptional
level. _Mol. Microbiol._ 32, 169–178 (1999). Article CAS PubMed Google Scholar * Lev, S. & Horwitz, B. A. A mitogen-activated protein kinase pathway modulates the expression of two
cellulase genes in _Cochliobolus heterostrophus_ during plant infection. _Plant Cell_ 15, 835–844 (2003). Article CAS PubMed PubMed Central Google Scholar * Zhang, G. et al. Functional
roles of laeA-like genes in fungal growth, cellulase activity, and secondary metabolism in _Pleurotus ostreatus_. _J. Fungi_ 8, https://doi.org/10.3390/jof8090902 (2022). * Cao, X. et al.
The role of hydrogen sulfide in cyclic nucleotide signaling. _Biochem. Pharmacol._ 149, 20–28 (2018). Article CAS PubMed Google Scholar * Kimura, Y. et al. Polysulfides are possible
H2S-derived signaling molecules in rat brain. _FASEB J._ 27, 2451–2457 (2013). Article CAS PubMed Google Scholar * Nagai, Y., Tsugane, M., Oka, J. & Kimura, H. Hydrogen sulfide
induces calcium waves in astrocytes. _FASEB J._ 18, 557–559 (2004). Article CAS PubMed Google Scholar * Castro-Piedras, I. & Perez-Zoghbi, J. F. Hydrogen sulphide inhibits Ca2+
release through InsP3 receptors and relaxes airway smooth muscle. _J. Physiol._ 591, 5999–6015 (2013). Article CAS PubMed PubMed Central Google Scholar * Jin, Z. et al. Hydrogen sulfide
interacting with abscisic acid in stomatal regulation responses to drought stress in _Arabidopsis_. _Plant Physiol. Biochem._ 62, 41–46 (2013). Article CAS PubMed Google Scholar *
Aroca, A., Yruela, I., Gotor, C. & Bassham, D. C. Persulfidation of ATG18a regulates autophagy under ER stress in _Arabidopsis_. _Proc. Natl Acad. Sci. USA_ 118,
https://doi.org/10.1073/pnas.2023604118 (2021). * Liu, Z. et al. The role of H2S in low temperature-induced cucurbitacin C increases in cucumber. _Plant Mol. Biol._ 99, 535–544 (2019).
Article CAS PubMed Google Scholar * Chen, S. et al. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. _Mol. Plant_ 13,
732–744 (2020). Article CAS PubMed Google Scholar * Jurado-Flores, A., Aroca, A., Romero, L. C. & Gotor, C. Sulfide promotes tolerance to drought through protein persulfidation in
_Arabidopsis_. _J. Exp. Bot._ 74, 4654–4669 (2023). Article CAS PubMed PubMed Central Google Scholar * Wang, R. Physiological implications of hydrogen sulfide: a whiff exploration that
blossomed. _Physiol. Rev._ 92, 791–896 (2012). Article CAS PubMed Google Scholar * Sen, N. et al. Hydrogen sulfide-linked sulfhydration of NF-κB mediates its antiapoptotic actions. _Mol.
Cell_ 45, 13–24 (2012). Article CAS PubMed PubMed Central Google Scholar * Wu, N., Siow, Y. L. & O, K. Ischemia/reperfusion reduces transcription factor Sp1-mediated cystathionine
beta-synthase expression in the kidney. _J. Biol. Chem._ 285, 18225–18233 (2010). Article CAS PubMed PubMed Central Google Scholar * Ge, Y., Konrad, M. A., Matherly, L. H. & Taub,
J. W. Transcriptional regulation of the human cystathionine beta-synthase-1b basal promoter: synergistic transactivation by transcription factors NF-Y and Sp1/Sp3. _Biochem. J._ 357, 97–105
(2001). Article CAS PubMed PubMed Central Google Scholar * Sun, Y. et al. A NAC transcription factor from ‘Sea Rice 86’ enhances salt tolerance by promoting hydrogen sulfide production
in rice seedlings. _Int. J. Mol. Sci._ 23, https://doi.org/10.3390/ijms23126435 (2022). * Ries, L. N., Beattie, S. R., Espeso, E. A., Cramer, R. A. & Goldman, G. H. Diverse regulation of
the CreA carbon catabolite repressor in _Aspergillus nidulans_. _Genetics_ 203, 335–352 (2016). Article CAS PubMed PubMed Central Google Scholar * Sun, J. & Glass, N. L.
Identification of the CRE-1 cellulolytic regulon in _Neurospora crassa_. _PloS One_ 6, e25654 (2011). Article CAS PubMed PubMed Central Google Scholar * Portnoy, T. et al. The CRE1
carbon catabolite repressor of the fungus _Trichoderma reesei_: a master regulator of carbon assimilation. _BMC Genom._ 12, 269 (2011). Article CAS Google Scholar * Chen, Y. et al. Carbon
catabolite repression governs diverse physiological processes and development in _Aspergillus nidulans_. _mBio_ 13, e0373421 (2021). Article PubMed Google Scholar * Fasoyin, O. E. et al.
Carbon catabolite repression gene creA regulates morphology, aflatoxin biosynthesis and virulence in _Aspergillus flavus_. _Fungal Genet. Biol._ 115, 41–51 (2018). Article CAS PubMed
Google Scholar * Hong, Y. et al. Carbon catabolite repressor MoCreA is required for the asexual development and pathogenicity of the rice blast fungus. _Fungal Genet. Biol._ 146, 103496
(2021). Article CAS PubMed Google Scholar * Luo, Z., Qin, Y., Pei, Y. & Keyhani, N. O. Ablation of the creA regulator results in amino acid toxicity, temperature sensitivity,
pleiotropic effects on cellular development and loss of virulence in the filamentous fungus _Beauveria bassiana_. _Environ. Microbiol._ 16, 1122–1136 (2014). Article CAS PubMed Google
Scholar * Beattie, S. R. et al. Filamentous fungal carbon catabolite repression supports metabolic plasticity and stress responses essential for disease progression. _PLoS Pathog._ 13,
e1006340 (2017). Article PubMed PubMed Central Google Scholar * Xu, W. et al. Mitochondrial pyruvate carrier regulates the lignocellulosic decomposition rate through metabolism in
_Ganoderma lucidum_. _FEMS Microbiol. Lett._ 368, https://doi.org/10.1093/femsle/fnab088 (2021). * Li, K. et al. Hydrogen Sulfide Regulates Glucose Uptake in Skeletal Muscles via
S-Sulfhydration of AMPK in Muscle Fiber Type-Dependent Way. _J Nutr._ 153, 2878–2892 (2023). Article CAS PubMed Google Scholar * Zhu, J. et al. Dual functions of AreA, a GATA
transcription factor, on influencing ganoderic acid biosynthesis in _Ganoderma lucidum_. _Environ. Microbiol._ 21, 4166–4179 (2019). Article CAS PubMed Google Scholar Download references
ACKNOWLEDGEMENTS This work was supported by: China Agriculture Research System (project number CARS20), the National Natural Science Foundation of China (project numbers 31972059 and
32272787), and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (project number KYCX220709). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Key Laboratory of
Agricultural Environmental Microbiology, Ministry of Agriculture and Rural Affairs; Department of Microbiology, College of Life Sciences, Nanjing Agricultural University, Nanjing, 210095,
Jiangsu, PR China Jiaolei Shangguan, Jinjin Qiao, He Liu, Lei Zhu, Xiaofei Han, Liang Shi, Jing Zhu, Rui Liu, Ang Ren & Mingwen Zhao Authors * Jiaolei Shangguan View author publications
You can also search for this author inPubMed Google Scholar * Jinjin Qiao View author publications You can also search for this author inPubMed Google Scholar * He Liu View author
publications You can also search for this author inPubMed Google Scholar * Lei Zhu View author publications You can also search for this author inPubMed Google Scholar * Xiaofei Han View
author publications You can also search for this author inPubMed Google Scholar * Liang Shi View author publications You can also search for this author inPubMed Google Scholar * Jing Zhu
View author publications You can also search for this author inPubMed Google Scholar * Rui Liu View author publications You can also search for this author inPubMed Google Scholar * Ang Ren
View author publications You can also search for this author inPubMed Google Scholar * Mingwen Zhao View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS J.S. and M.W.Z. designed the study. J.S., J.Q., H.L., L.Z., and X.H. carried out experiments and analyzed data. L.S., J.Z., R.L., A.R., and M.W.Z. provided supervisor
oversight. J.S. wrote the manuscript. All authors gave input and approved the manuscript. CORRESPONDING AUTHOR Correspondence to Mingwen Zhao. ETHICS DECLARATIONS COMPETING INTERESTS The
authors declare no competing interest. PEER REVIEW PEER REVIEW INFORMATION _Communications Biology_ thanks Pedro Gonçalves and the other, anonymous, reviewer(s) for their contribution to the
peer review of this work. Primary Handling Editor: David Favero. A peer review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION PEER REVIEW FILE SUPPLEMENTARY INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES
SUPPLEMENTARY DATA 1 REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing,
adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons
licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a
credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted
use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT
THIS ARTICLE CITE THIS ARTICLE Shangguan, J., Qiao, J., Liu, H. _et al._ The CBS/H2S signalling pathway regulated by the carbon repressor CreA promotes cellulose utilization in _Ganoderma
lucidum_. _Commun Biol_ 7, 466 (2024). https://doi.org/10.1038/s42003-024-06180-y Download citation * Received: 17 October 2023 * Accepted: 10 April 2024 * Published: 17 April 2024 * DOI:
https://doi.org/10.1038/s42003-024-06180-y SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Susannah townsend calls time on her international hockey career | england hockeyAfter playing 188 games across a 13-year career, Susannah Townsend has retired from international hockey. A double Olymp...
Video 'Big Little Lies' star Laura Dern teases possible return to 'Jurassic Park' - ABC NewsABC NewsVideoLiveShowsShopStream onLive UpdatesLive UpdatesTrump 2nd term Ukraine drone attack Trump tariffs Boulder att...
Do people still celebrate their name days in france?IT IS A FRENCH TRADITION TO SEND PEOPLE GOOD WISHES ON THEIR ‘NAME DAY’. WE LOOK AT WHERE THIS CAME FROM AND WHETHER THE...
Sniffing out genes' role in our senses of taste and smellPHILADELPHIA — Armed with jellybeans, Q-tips dipped in bitter liquid and an assortment of rare truffles, Marcie Pelchat ...
New approaches to quantifying the spread of infectionKEY POINTS * The last decade has seen considerable advances in statistical, mathematical and computational techniques th...
Latests News
The cbs/h2s signalling pathway regulated by the carbon repressor crea promotes cellulose utilization in ganoderma lucidumABSTRACT Cellulose is an important abundant renewable resource on Earth, and the microbial cellulose utilization mechani...
404: This page could not be foundआरएसएसविज्ञापन र॓टहमार॓ साथ काम करेंहमारे बारे मेंसंपर्क करेंगोपनीयतासाइट जानकारीAdvertise with usAbout usCareers Privac...
Forecast cloudy for comprehensive immigration reformI’ve noticed a shift in the last week among comprehensive immigration reform advocates from hopeful to downright angry a...
Base editing boosts hemoglobin in sickle cell diseaseYou have full access to this article via your institution. Download PDF Beam Therapeutics’ early clinical data on its ba...
Wearable cuffless blood pressure tracking: when will they be good enough?ABSTRACT Wearable health monitoring is a multibillion-dollar industry. But the holy grail is probably getting it right f...