Madr-net: multi-level attention dilated residual neural network for segmentation of medical images
Madr-net: multi-level attention dilated residual neural network for segmentation of medical images"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Medical image segmentation has made a significant contribution towards delivering affordable healthcare by facilitating the automatic identification of anatomical structures and
other regions of interest. Although convolution neural networks have become prominent in the field of medical image segmentation, they suffer from certain limitations. In this study, we
present a reliable framework for producing performant outcomes for the segmentation of pathological structures of 2D medical images. Our framework consists of a novel deep learning
architecture, called deep multi-level attention dilated residual neural network (MADR-Net), designed to improve the performance of medical image segmentation. MADR-Net uses a U-Net
encoder/decoder backbone in combination with multi-level residual blocks and atrous pyramid scene parsing pooling. To improve the segmentation results, channel-spatial attention blocks were
added in the skip connection to capture both the global and local features and superseded the bottleneck layer with an ASPP block. Furthermore, we introduce a hybrid loss function that has
an excellent convergence property and enhances the performance of the medical image segmentation task. We extensively validated the proposed MADR-Net on four typical yet challenging medical
image segmentation tasks: (1) Left ventricle, left atrium, and myocardial wall segmentation from Echocardiogram images in the CAMUS dataset, (2) Skin cancer segmentation from dermoscopy
images in ISIC 2017 dataset, (3) Electron microscopy in FIB-SEM dataset, and (4) Fluid attenuated inversion recovery abnormality from MR images in LGG segmentation dataset. The proposed
algorithm yielded significant results when compared to state-of-the-art architectures such as U-Net, Residual U-Net, and Attention U-Net. The proposed MADR-Net consistently outperformed the
classical U-Net by 5.43%, 3.43%, and 3.92% relative improvement in terms of dice coefficient, respectively, for electron microscopy, dermoscopy, and MRI. The experimental results demonstrate
superior performance on single and multi-class datasets and that the proposed MADR-Net can be utilized as a baseline for the assessment of cross-dataset and segmentation tasks. SIMILAR
CONTENT BEING VIEWED BY OTHERS MULTI-LEVEL DILATED RESIDUAL NETWORK FOR BIOMEDICAL IMAGE SEGMENTATION Article Open access 08 July 2021 CMM-NET: CONTEXTUAL MULTI-SCALE MULTI-LEVEL NETWORK FOR
EFFICIENT BIOMEDICAL IMAGE SEGMENTATION Article Open access 13 May 2021 A NOVEL MEDICAL IMAGE SEGMENTATION APPROACH BY USING MULTI-BRANCH SEGMENTATION NETWORK BASED ON LOCAL AND GLOBAL
INFORMATION SYNCHRONOUS LEARNING Article Open access 25 April 2023 INTRODUCTION Biomedical imaging is an indispensable tool for applications such as localization of pathology, disease
diagnosis, treatment planning, and disease management. The most commonly used non-interventional/invasive diagnostic mapping includes computed tomography, magnetic resonance imaging (MRI),
digital mammography, fundus imaging, and other imaging modalities. These medical imaging tools offer the first line of choice due to their low cost, real-time functionality, and portability.
Image formulation and reconstruction along with image processing and analysis are the two components of medical imaging1. Image formulation and reconstruction involve the process through
which 2D and 3D images are typically formed from the projection data of an object. On the other hand, image processing involves enhancing the image properties to facilitate object
identification and classification. Ease of image acquisition has paved the way for producing high-resolution images at extremely low cost2. Computer-aided diagnosis for delineation of
pathological structures is becoming an important tool for clinicians. Doctors use medical images to judge the condition of patients for clinical diagnosis. Medical image segmentation plays a
crucial role in computer-aided diagnosis and intends to visualize the changes in the pathological or anatomical structure of the images. The segmentation techniques can be grouped into
three different categories: (1) Manual segmentation, (2) Semi-automatic segmentation, and (3) Fully automatic segmentation. Manual segmentation is the initial step for determining the region
of interest (ROI) and for precisely annotating the boundaries. Further, semi-automation techniques involve a user interface for initial ROI segmentation from the entire image. The fully
automatic segmentation technique is based on supervised learning approaches, and they do not require any user interaction. Currently, medical image segmentation is semi-automatic and suffers
from various complications3: (1) The whole process needs to be carried out only by an experienced clinician, (2) The annotations will always include inter and intra-observer variability,
and (3) This is a cumbersome and error-prone process that must be done for each patient individually. Consequently, automatic segmentation approaches have been developed to address these
issues and facilitate higher patient throughput and lower inter-user discrepancy. However, automatic segmentation also suffers from a number of issues: (1) Low signal-to-noise ratio, (2)
Poor contrast between the myocardium and blood pool, (3) Motion artifacts of the heart structures across patients and pathologies, (4) Brightness inhomogeneities, and (5) Low spatial and
temporal resolutions4. Acknowledging the relevance and importance of segmentation tasks mentioned above, automation of this activity has been a major study topic of research in recent
decades5,6,7,8,9,10,11,12,13,14. Prior to the widespread application of deep learning, researchers utilized model-driven strategies (active contours, level sets, deformable models, and
statistical shape models) for medical image segmentation, which typically require manual interventions. However, the emerging interest in computer-aided diagnosis (CAD) models aims to
automate these processes for increased efficiency and accuracy. To achieve this objective, researchers proposed the encoder-decoder structure such as a fully convolutional network (FCN)15,
U-Net16, and Deeplab17 which effectively automate image segmentation tasks. Badrinarayanan et al.18 proposed a deep convolutional encoder-decoder architecture that allows to perform
pixel-level semantic segmentation. Ronneberger et al.16 extended Long et al.'s FCN15, by introducing the classical U-Net architecture, known for its end-to-end training ability and
widespread application in the field of biomedical image segmentation. Although U-Net is effective, it has limitations such as loss of spatial information and difficulty in handling image
variations. Several variations of U-Net architectures, such as Attention U-Net19, Residual U-Net20, Multi Residual U-Net21, V-Net22, R2 U-Net23 and U2Net24, have been proposed in the
literature. In this paper, while analyzing the strengths of U-Net architecture, we delicately examined the network architecture to identify a promising scope for future development. To
extract features from different scales and sizes, we suggest replacing the convolution blocks of the classic U-Net with multi-dilated residual convolution blocks of rate d = 1, 3, 5, and 11
in this study. Embedding the ASPP module at the bottleneck position provides the network ability to efficiently capture multi-scale contextual information. Finally, the utilization of the
channel-spatial attention mechanism played a pivotal role in extracting both channel-wise and spatial information, while suppressing noise and irrelevant features that are crucial for
analysis. In view of the above, we designed a novel deep learning architecture called deep multi-level attention dilated residual neural network (MADR-Net), that incorporates a class-spatial
attention mechanism-driven decoder and inculcates the properties of dilated residual neural networks and atrous spatial pyramid pooling. Unlike the classic U-Net, MADR-NET extracts the
features at different scales by integrating multiple dilated convolution modules. This process increases segmentation accuracy and captures multi-scale information.To extract the features at
multiple scales and rich contextual details, atrous spatial pyramid pooling (ASPP) module with various dilation rates is used in the bottleneck layer of the encoder-decoder architecture.
Further, channel-spatial attention modules are employed in skin pathways to suppress unnecessary areas in an input image while emphasizing important characteristics for a particular task. In
this study, a hybrid loss function with a combination of cross-entropy, dice loss, and focal Tversky loss was introduced to enhance the models’ ability to quantify discrepancies between
predicted and ground truth in medical image segmentation tasks. To validate the performance of the proposed MADR-Net architecture, four challenging clinical segmentation problems were
addressed, namely echocardiogram, dermoscopy, electron microscopy, and MR images, demonstrating its efficacy across diverse medical imaging modalities. To validate the performance of the
proposed architecture, we adopted three different architectures which include U-Net, Res Net, and attention U-Net. The main contributions of this proposed work are as follows: * MADR-Net is
based on the classical U-Net architecture of several biomedical imaging datasets. To overcome the loss of spatial information in U-NET, the convolutional block of U-Net is replaced by the
multi-level dilated residual network. * Channel-spatial attention modules are implemented to extract both shallow and deep feature maps, which increases the focus on the area of interest of
target segmentation, resulting in the finest precision across all experiments. * Based on our findings, hybrid loss function, speeds up the convergence and improves the performance of the
segmentation task. * MADR-Net has demonstrated its potential as a feasible and generalizable solution across various image modalities, including electron microscopy, dermoscopy,
echocardiogram, and MRI, through its ability to segment medical tasks. * The robustness of the proposed MADR-Net architecture is compared to recent state-of-the-art architectures such as
U-Net, Attention U-Net, Multi residual U-Net, SU-Net, CNLU-Net and Res U-Net architectures. In this paper, "Prior art" section discusses the related traditional and deep learning
approaches used for the segmentation of various image modalities. "Network architecture" section highlights the network architectures, its components, and methodology.
"Experimental setup" section presents the results and discussion in terms of ablation and comparison study. Finally, the conclusions of the study are presented in "Results and
discussion" section. PRIOR ART TRADITIONAL MACHINE LEARNING TECHNIQUES Traditional artificial intelligence approaches are based on computer-aided diagnosis systems that extract
features from spatial, temporal, and morphological regions. However, the effectiveness of extracting these features is a real challenge in medical images due to their susceptibility to noise
and motion artifacts. Medical image segmentation often depends on Level set25, multilevel thresholding26, Active shape models27, Active contour model28, Active appearance models29,
Bottom-up method30, and Database-guided31. Bhandari et al.32 introduced different objective functions using cuckoo search to solve image segmentation through multilevel thresholding. Cheng
and Wang33 presented homogram thresholding and region-based merging for color image segmentation. Ming-Ni Wu et al.34 proposed a color-based segmentation to track brain tumors using k-means
clustering. Although several algorithms have been described and are successful in certain situations, segmentation of medical images remains one of the most difficult subjects in computer
vision due to the complexity of feature representation. DEEP LEARNING TECHNIQUES Recently, deep learning approaches have demonstrated promising results in segmentation tasks due to their
capacity to learn complicated characteristics from data35. Convolutional neural networks are commonly utilized for medical image processing applications such as localization, segmentation,
and classification. Plain convolutional neural networks (CNN) have been extended to numerous networks, including Deep feedforward neural networks36, Deep long short-term memory (LSTM)
architecture37, Inception CNN architecture38, Deep belief network (DBN)39, and Deep generative adversarial architecture (DGAN)40. Based on these, Fully convolutional network (FCN)15 and
U-Net16 were proposed for semantic segmentation as they have exhibited remarkable performance. FCN architecture is trained end-to-end for pixel-wise prediction. U-Net architecture consists
of two paths: encoder and decoder path. In the encoder or analysis part, deep features are learned and in the decoder part, segmentation is performed based on the learned features. These
architectures have been applied to several 2D and 3D medical images for automated medical image assessment. Popular medical image segmentation tasks include brain tumor segmentation from
magnetic resonance imaging, skin lesion segmentation from dermoscopy images, segmentation of the left ventricle from echocardiography images, and segmentation of the hippocampus region from
electron microscopy images. Table 1 shows the visual comparison of the proposed MADR-Net with the baseline approaches with a detailed description of the encoder, decoder, bottleneck, and
skip connection layers. * (i) _Segmentation of left ventricle, left atrium, and myocardial wall from Echocardiogram images:_ Sarah Leclerc et al.5 introduced the largest publicly available
dataset for 2D echocardiogram assessment. To get the best possible results on this dataset, a modified U-Net architecture was proposed, and the experts reached excellent agreement for the
estimation of left ventricle end-diastolic (ED) and end-systolic (ES) with dice scores of 0.939 ± 0.043 and 0.916 ± 0.061, respectively. A lack of training data, low signal-to-noise ratio,
and substantial variability among perspectives for collaborative learning are some of the limitations of echocardiogram data. To address these issues, Li et al.41 developed a multiview
recurrent aggregation network (MV-RAN) for full cardiac cycle analysis. The MV-RAN architecture produced an average dice score of 0.92 ± 0.04 for the segmentation of the left ventricle. Liu
et al.42 presented a deep learning model for the automatic segmentation of a 2D echocardiogram based on a deep pyramid local attention neural network. The correlation between the actual left
ventricle ejection fraction and predicted left ventricle ejection fraction was 0.883 and 0.869, respectively for two different datasets, whereas the corresponding dice scores were 0.951 and
0.943 for ED and ES frames, respectively. Ali et al.43 proposed a fast and automatic deep learning framework by fusing Res Net and U-Net to enhance the segmentation results. Guo et al.44
presented a fusion of low-level and high-level features based on a spatial attention module along with a hybrid loss function. * (ii) _Segmentation of skin lesion from dermoscopy images_:
The ISIC 2017 skin lesion analysis challenge was arranged by Bi et al.45, and the multiscale residual network took first place with an average Jaccard index of 79.40%. The segmentation of
skin lesions remains a difficult process due to poor contrast, blurred boundaries, and varying sizes of cancer patches. To alleviate these drawbacks, Zhang et al.46 developed a deep
supervised multi-scale network for the segmentation of skin lesions by designing a multi-scale connection block along with shallow and deep layers. The experiments were carried out on two
different datasets and an average dice score of 87.5% and Jaccard index of 78.5% were reported. For the segmentation of skin lesions, Wei et al.47 suggested a novel attention-dense U-Net
with adversarial training, which yielded a dice score of 0.8786 for the ISIC2017 dataset. Hasan et al.48 proposed a novel semantic segmentation network for robust skin lesion segmentation
using depth-wise separable convolution instead of normal convolution to reduce the parameters of the network. To capture context characteristics and higher semantic feature information, Tang
et al.49 presented a separable U-Net with stochastic weight averaging with an average dice coefficient of 86.93% and a Jaccard index of 79.26%. * (iii) _Brain tumor segmentation from
magnetic resonance images_: Zhao et al.50 proposed a combination of a fully convolutional neural network along with conditional random fields (CRF) to segment the brain tumor. With 110 BRATS
2015 test cases, Kamnitsas et al.51 developed a dual pathway architecture built at multiple scales and obtained an average dice score of 0.847 for the automated segmentation of brain
tumors. Havaei et al.52 explored different architectures based on CNN to segment glioblastomas i.e., low and high-grade MR images. To avoid overfitting in neural networks, Pereira et al.53
developed dropout, ReLu, and tiny convolution layers to discriminate between high and low-grade glioma images. In 2019, Wadhwa et al,54 observed that the combination of CRF with FCNN and CRF
with DeepMedic is more successful in the segmentation of brain tumors. * (iv) _Segmentation of mitochondria from electron microscopy images_: Automatic segmentation of mitochondria and
reconstruction in electron-microscopy images has proven to be a challenging task due to the variety of mitochondrial structures. Xiao et al.55 proposed a method for 3D mitochondria
segmentation based on residual convolutional and highly supervised networks to overcome this problem. Manca et al.56 presented an automatic method for segmenting intracellular compartments
such as mitochondria and endolysosomes. The authors compared the proposed algorithm with U-Net, V-Net, and DeepMedic algorithms and obtained dice scores of 0.855, 0.898, and 0.867,
respectively. Oztel et al.57 reported encouraging findings for the autonomous segmentation of mitochondria in brain tissue using a deep convolutional neural network technique. Chacon et
al.19 introduced a domain adaptation approach that relies on two coupled U-Net that share weights as well as a differentiable loss function that approximates the Jaccard index. NETWORK
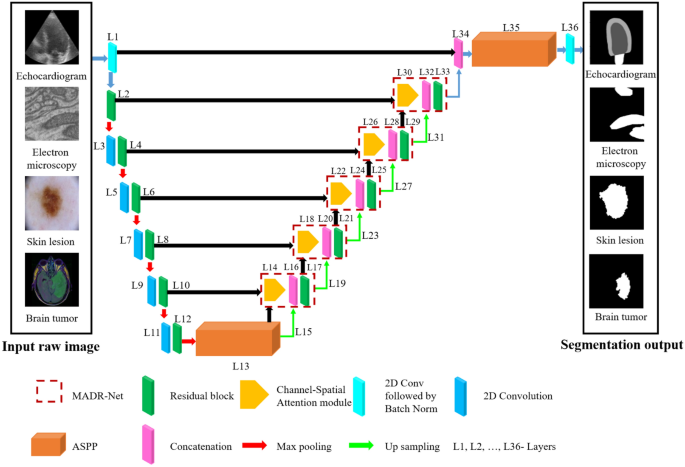
ARCHITECTURE In view of the shortcomings of the fundamental architecture, three important design enhancements were made. Fig. 1 depicts an overview of the proposed architecture. Inspired by
U-Net16, Residual U-Net58, and ASPP17 architectures, a novel segmentation model named MADR-Net has been developed. The proposed MADR-Net architecture has 37 layers and takes advantage of the
channel-spatial attention mechanism, ASPP, and multi-scale residual blocks. The multi-scale residual blocks help in tackling the vanishing gradient problem and build deeper neural networks
to solve the degradation problem in each of the encoders. Each encoder block consists of \(3\times 3\) convolution blocks followed by multi-scale residual blocks and each multi-scale
residual block has four parallel branches with a set of two stacked convolution layers such as batch normalization, ReLU, and 2D convolution layer. The outputs from the convolutional layers
are added instead of concatenation to make the training earlier. An ASPP operator with four equal (1, 6, 12, 18) partitions has been used to bridge the encoder and decoder paths. In the
decoder path, upsampling along with 2D convolution followed by batch normalization helps to improve the resolution of the convolutional feature59. The output of the decoder block is sent
through the ASPP block and depending on the segmentation task \(1\times 1\) convolution with sigmoidal or softmax activation was applied. The different paths of the model notably the
encoder, bottleneck, and decoder paths have been delineated as follows. ENCODER PATH Deep networks are naturally integrated with different levels of features (low/mid/high) and these
features can be enriched by the depth or number of stacked layers. Driven by the significance of depth, the problem of vanishing grading could hamper the training process, and degradation
problems may occur. To overcome these problems, He et al.60 introduced a deep residual learning framework that directly fits a desired underlying mapping instead of hoping for each stacked
layer. Moreover, residual blocks are known to provide a deeper network and these connections facilitate model learning with reference to the input layer instead of learning from an
unreferenced function. The current U-Net architecture is a symmetric model and has only a few layers. We have replaced the basic U-Net architecture with the Multi-Dilated Residual blocks as
shown in Fig. 2a. These residual blocks further contribute to feature propagation at both encoder and decoder paths. The general form of residual block can be expressed as $$y_{N} = W_{s}
\left( {x_{N} } \right) + {\mathcal{F}}\left( {x_{N} ,\left\{ {W_{N} } \right\}} \right)$$ (1) $${x}_{N+1}=f({y}_{N})$$ (2) Where \({x}_{N}\) and \({x}_{N+1}\) are the input and output with
the Nth block respectively. \({\mathcal{F}}\) and f represent the residual mapping function and ReLU function. \({W}_{s}\left({x}_{N}\right)={x}_{N}\) which is an identity mapping. The
performance of the residual block of Pre-ResNets is computed as follows $$x_{N + 1} = W_{s} \left( {x_{N} } \right) + {\mathcal{F}}\left( {x_{N} ,\left\{ {W_{N} } \right\}} \right)$$ (3) In
this study, the encoder path consists of 12 layers, which start with a 1 \(\times 1\) convolution layer to avoid information loss, followed by a Multi-Dilated Residual block with an input
size of \(112\times 112\times 3\). Each multi-dilated residual block consists of \(3\times 3\) parallel dilated convolutions at dilation rates of 1, 3, 5, and 11 along with the residual
connection to extract features at different resolutions. Fig. 3 illustrates the 2D convolution with a spatial size of \(3\times 3\) and different dilation rates (d = 1, 3, 5). In dilated
convolution, the kernel with a \(k\times k\) filter is amplified to \(k+(k-1)(d-1)\). Normal convolution gets a \(3\times 3\) receptive field, whereas dilated convolution with a dilation
rate of 3 and 5 provides \(5\times 5\) and \(11\times 11\) receptive fields. Therefore, while normal convolution and dilated convolution have the same number of parameters, dilated
convolution has a larger receptive field. BOTTLENECK In the proposed approach, the atrous spatial pyramid pooling (ASPP)17 acts as the intermediary area that bridges the encoder and decoder
paths. The ASPP module was developed to overcome the requirement of the constant size of the input image and resample features at multiple scales. The ASPP layer enlarges the field of view
of the filters by capturing multiscale information precisely, as shown in Fig. 2b. In this study, multi-scale information can be captured through the ASPP module that consists of (1, 1)
convolution followed by (3, 3) convolutions with different dilated rates (d = 6, 12, and 18) and a parallel max-pooling. The output of the bottleneck layers is sent through two separate
paths: the convolutional transpose layer and the attention gate layer. DECODER PATH The decoder path consists of 23 layers, each of which has a multi-dilated residual network, 2 \(\times\) 2
convolutional transpose layer, skip connection, and Channel-spatial attention mechanism. Each decoder layer is coupled to the encoder layer by combining (L14, L10), (L18, L8), (L22, L6),
(L26, L4), and (L30, L2) layers. Class-spatial attention module receives the output from the encoder layer and the preceding layer9. Initially, both average pooling (\({F}_{avg}^{C}\)) and
max pooling (\({F}_{max}^{C}\)) operations are used to gather aggregate spatial information from the feature maps \(({F}_{1}\)). These spatial context descriptors are then forwarded to a
shared network which consists of a multi-layer perceptron with one hidden layer to produce a channel attention map \({W}_{C}\in {\mathbb{R}}^{C\times 1\times 1}\). The channel attention is
computed as: $${W}_{C}(F)=\sigma \left({W}_{1}\left({W}_{0}\left({F}_{avg}^{C}\right)\right)+{W}_{1}\left({W}_{0}\left({F}_{max}^{C}\right)\right)\right)$$ (4) Where \({W}_{0}\in
{\mathbb{R}}^{C/r\times C}\) and \({W}_{C}\in {\mathbb{R}}^{C\times C/r}\) are the MLP weights that are shared for both inputs and _r_ is the reduction ratio. In the spatial attention module
(SAM), average pooled features (\({F}_{avg}^{S}\)) and max pooled features (\({F}_{max}^{S}\)) across the channel are concatenated and convolved by a standard convolution layer
(\({f}_{7\times 7}\)) with the filter size of \(7\times 7\) producing a spatial attention map \({W}_{S}\in {\mathbb{R}}^{1\times H\times W}\). The spatial attention is computed as:
$${W}_{S}(F)=\sigma \left({f}_{7\times 7}\left(\left[{F}_{avg}^{S};{F}_{max}^{S}\right]\right)\right)$$ (5) $${F}_{1}={W}_{C}(F)\otimes F$$ (6) $${F}_{2}={W}_{S}({F}_{1})\otimes {F}_{1}$$
(7) Where \(F\in {\mathbb{R}}^{C\times H\times W}\) is the intermediate feature and \(\otimes\) represents element-wise multiplication. Finally, the attention block's output is
concatenated with the preceding layer's upsampled output. Fig. 4, illustrates two sequential sub-modules: (1) channel attention module and (2) spatial attention module. At each and
every convolutional block in deep networks, the intermediate feature map is adaptively improved by the channel-spatial attention module. The output is given to the residual block after
concatenation, which is the same as the encoder path. Finally, the output of the multi-dilated residual block is combined with L1 and passed through ASPP architecture. EXPERIMENTAL SETUP To
evaluate the performance of the proposed deep multi-level attention dilated residual neural network (MADR-Net) architecture, we have tested it on four distinct medical image datasets. This
section discusses the loss function used in the training process, dataset, and preprocessing steps involved in MADR-Net. Finally, the results of MADR-Net are compared with other
state-of-the-art techniques on the different medical image datasets. DATASET The effectiveness of the proposed MADR-Net architecture is evaluated with four publicly available datasets from
different image modalities. Due to large variations in image sizes ranging from 256 \(\times\) 256 to 1022 \(\times\) 767 pixels, we rescalled all the data to 128 \(\times\) 128 pixels. All
datasets are normalized and to reduce the overfitting problem, different augmentation techniques were employed. Table 2 provides a full overview of the biomedical imaging dataset utilized in
the proposed investigation. * Cardiac Acquisitions for Multi-Structure Ultrasound Segmentation (CAMUS)5 consist of a two-dimensional apical two-chamber and four-chamber view sequence. *
Dermoscopy images were acquired from the Medical Image Computing and Computer-Aided Intervention (MICCAI)61 conference hosted by the International Skin Imaging Collaboration (ISIC) in 2017
for analysis of skin lesions. * FIB-SEM62 dataset consists of serial section transmission electron microscopy (SSTEM) images acquired from the hippocampus region of the brain. * Brain MR
images with manual ground truth of fluid-attenuated inversion recovery (FLAIR) abnormalities were acquired from The Cancer Imaging Archive (TCIA)63. EVALUATION METRICS The performance of the
segmentation model can be reliably evaluated in terms of dice similarity coefficient (DSC), Jaccard index or intersection over union (IoU), accuracy, sensitivity, and specificity. The
disjoint sets are defined as follows: True positive (TP) set as \(TP=GT\cap PR\), True negative (TN) set as \(TN=\overline{GT }\cap \overline{PR }\), False positive (FP) set as
\(FP=\overline{GT }\cap PR\) and False negative (FN) set as \(FN=GT\cap \overline{PR }\). In medical segmentation, the region of interest will be too small compared to the entire image, so
TP will be low, and the background or non-infected region will be represented as TN64. This may lead to misleading performance, and so to overcome the class imbalance, it is necessary to
focus on DSC and IoU metrics that robustly reflect the performance of the model. $$DSC=\frac{2|GT\cap PR|}{\left|GT\right|+|PR|}$$ (8) $$IoU=\frac{|GT\cap PR|}{|GT\cup PR|}$$ (9) In the
above equations, PR and GT are the predicted region and their corresponding ground truth. The measurements were graded from 0 (lowest) to 1 (highest). The performance metrics TN, TP, FN, and
FP were used to evaluate accuracy, precision, and recall. $$Accuracy=\frac{TP+TN}{(TP+TN+FP+FN)}$$ (10) $$Precision=\frac{TP}{(TP+FP)}$$ (11) $$Recall=\frac{TP}{(TP+FN)}$$ (12) LOSS
FUNCTION In our designed model, we used the combination of binary/categorical cross-entropy, dice loss, and focal Tversky loss to train the model. The binary cross-entropy
(\({{\ell}}_{BCE}\))65 loss function is computed as $${{\ell}}_{BCE}=-\frac{1}{N}\sum_{i=1}^{N}{y}_{i}.log{\widehat{y}}_{i}+(1-{y}_{i})\text{log}(1-{\widehat{y}}_{i})$$ (13) where
\({\widehat{y}}_{i}\) is the _i_th scale value in the model output, \({y}_{i}\) is the corresponding target value and N is the output size. For the multi-class problem, the categorical
cross-entropy loss function (\({{\ell}}_{CCE}\)) is computed as follows: $${{\ell}}_{CCE}=-\frac{1}{N}\sum_{m=1}^{M}\sum_{i=1}^{N}{y}_{i}^{m}.log{\widehat{y}}_{i,m}$$ (14) where
\({\widehat{y}}_{i,m}\) is a matrix of predicted values for each class, where _i_ and _m_ iterate over all pixels and classes, respectively. Cross entropy loss is predicted by minimizing
pixel-wise error, which results in poor quality segmentation of smaller ROI. Apart from the cross-entropy loss function, the Sorensen-Dice index66 was used for evaluating segmentation
accuracy. The DSC can be defined in terms of the per-pixel classification of TP, FP, and FN which is given in Eq. (4). The dice loss (\({{\ell}}_{DSC}\)) can be defined as:
$${{\ell}}_{DSC}=1-DSC$$ (15) On the other hand, the dice loss gradient is intrinsically unstable with unbalanced class data with a small denominator67. Focal Tversky (\({{\ell}}_{FT}\))68
loss is defined as: $${{\ell}}_{FT}=\sum_{m=1}^{M}{\left(1-\frac{\sum_{i=1}^{N}{p}_{0i}{q}_{0i}}{{\sum }_{i=1}^{N}{p}_{0i}{q}_{0i}+\alpha \sum_{i=1}^{N}{p}_{0i}{q}_{1i}+\beta
\sum_{i=1}^{N}{p}_{1i}{q}_{0i}}\right)}^{\frac{1}{\gamma }}$$ (16) where \({p}_{0i}\) and \({p}_{1i}\) are the probability of pixel _i_ belonging to the foreground class and background
class, respectively. \({q}_{0i}\) takes the value of 1 for the foreground and 0 for the background, whereas \({q}_{1i}\) is 1 for the background and 0 for the foreground. Here, we have
chosen \(\alpha =0.7 \text{and} \beta =0.3\) to improve recall in the case of large class imbalances. To overcome the segmentation of small ROI, the focal Tversky loss function with \(\gamma
=1.33\) was used to control the background and hard ROI. The hybrid loss adapts cross-entropy loss in combination with dice loss and focal Tversky losses to handle class imbalance. RUN TIME
Experiments were performed in a server with an Intel(R) Silver(R) 4210 CPU on 2.19 GHz and 128GB RAM. The training time of the MADR-Net with the best performance was 14 hrs and 4 seconds
per image for testing. The proposed architecture is optimized by Adam optimizer and started the training with a batch size of 16. The learning rate of the proposed algorithm is set to 1⨯10−3
which slowed down the convergence rate. The size of the images in each dataset is different, so the images are resized before feeding them to the model. We have utilized 80% data for
training, 10% of the dataset for testing, and the remaining 10% for validation. All the models were trained for 500 epochs at a reduced learning rate to create a more generic model. RESULTS
AND DISCUSSION To validate the performance of the proposed MADR-Net architecture, we trained, validated, and tested the model on four different publicly available datasets (FIB-SEM, ISIC
2017, LGG, and CAMUS). In this section, the proposed architecture is compared with other baseline architectures such as U-Net16, Attention U-Net20, and Residual U-Net58 for the different
segmentation tasks. ABLATION STUDY ON THE PROPOSED MADR-NET ON BINARY SEGMENTATION The effectiveness of the proposed model is evaluated by binary segmentation of the infected part from the
non-infected tissue. The influence of class distribution on the performance of semantic segmentation has been investigated. The segmentation performance of the proposed MADR-Net model is
illustrated in Tables 3 and 4. Along with the performance measures, training times for all the architectures were noted for 500 epochs. As compared to other networks such as U-Net,
Attention, and Residual U-Net, our proposed MADR-Net achieves an average improvement of 4.16%, 7.1%, and 1.75% (in terms of DSC), and 5.43%, 8.73% and 2.38% (on Jaccard) respectively for the
FIB-SEM dataset. For ISIC 2017 dataset, the dice scores for U-Net, Attention U-Net, Residual U-Net, and MADR-Net are 87.08%, 86.64%, 88.14%, and 89.46% respectively. It can be seen that the
dice score for MADR-Net outperforms the U-Net, attention U-Net, and residual U-Net by 2.74%, 3.43%, and 9.6%, respectively for the LGG dataset. Furthermore, the proposed algorithm offers
comparable accuracy, precision, and recall on the test set of FIB-SEM, ISIC 2017, and LGG datasets for binary segmentation. ABLATION STUDY ON THE PROPOSED MADR-NET ON MULTI-CLASS
SEGMENTATION Left ventricle endocardium (LV endo), left atrium (LA), and left ventricle epicardium (LV epi) gathered under one roof in the segmentation process and the proposed architecture
was used to semantically segment all the three classes. Table 4 shows the comparison of the proposed MADR-Net with other state-of-the-art architectures on the CAMUS dataset for multi-class
segmentation. The proposed algorithm yields a LV epi dice score of 96.20% and outperforms baseline architectures by 1.24%, 1.04%, and 1.66% for U-Net, Attention U-Net, and Residual U-Net,
respectively. Similarly, the proposed architecture yields an average improvement of 0.16%, 0.31%, and 0.1785% (in terms of Endo DSC) for U-Net, Attention U-Net, and Residual U-Net. However,
the maximum LA DSC is achieved by U-Net (90.07%) when compared to the proposed MADR-Net (89.90%). The maximum accuracy of 95.70% and minimum accuracy of 94.53% are obtained for the MADR-Net
and Residual U-Net datasets, respectively. Precision and recall of 95.72% and 95.21% are obtained for the multiclass segmentation task. PERFORMANCE AND PARAMETER COMPARISON OF DIFFERENT
ARCHITECTURES The size of the input image is 128 × 128 pixels and the state-of-the-art methods (U-Net, V-Net, Att U-Net, U-Net ++, R3 U-Net, and Res U-Net) are evaluated for different
segmentation tasks with the following structure: 1 \(\to\) 32 \(\to\) 64 \(\to\) 128 \(\to\) 256 \(\to\) 512 \(\to\) 1024 \(\to\) 512 \(\to\) 256 \(\to\) 128 \(\to\) 64 \(\to\) 32 \(\to\) 1.
Fig. 5 illustrates the number of training parameters and performance measures (DSC) for each segmentation technique on ISIC 2017 dataset. To provide a fair comparison, all the
state-of-the-art techniques are replicated using the original code provided in their article, while maintaining the same preprocessing and training environment. Table 1S reports the
parameter, inference time, and segmentation results obtained from ISIC 2017 dataset. To evaluate the effectiveness of the proposed MADR-Net method, we conducted a comparison with
state-of-the-art techniques based on the number of parameters and DSC using 128 × 128 input image. However, the training time per each epoch is comparatively large depending upon the
dataset. The training time for the proposed MADR-Net model took approximately 7 hours, while the testing time per image was 0.91 seconds. In addition, the proposed MADR-Net technique
required a moderate number of trainable parameters which is 56M compared to 66M, 92M and 75M in the cases of V-Net, R2U-Net and Res U-Net architectures, respectively (see the supplementary
Table 1S for further details). Overall, the proposed MADR-Net architecture has the potential to be viable in routine clinical settings due to its remarkable efficiency and superior
segmentation performance. VISUAL INSPECTION OF FEATURE MAPS Using ablation experiments and quantitative evaluations to demonstrate performance differences may not be adequate to properly
grasp the advantage and behavior of the proposed MADR-Net model. Tables 3 and 4 show the superior performance of the proposed architecture. Fig. 6 depicts the segmentation results of skin
lesion, electron microscopy, and MRI with different baseline architectures. The study also focused on the comparison of different architectures in terms of convergence. Figs. 6 and 7
visualize the segmentation output of single-class and multi-class segmentation with different baseline architectures. Fig. 7a and b illustrate the raw input images and their corresponding
ground-truth masks of the echocardiogram image with ED and ES frames. Fig. 7c–f represents the predicted segmentation masks of U-Net, Attention U-Net, and Residual U-Net. Fig. 8 depicts the
mean DSC value for each of the networks. It can be observed that, while the majority of the baseline architecture generates results that are similar, the convergence of the proposed MADR-Net
is quicker than other architectures for the segmentation of the various datasets. This is due to the interaction of multi-scale residual blocks and ASPP networks, which allows the MADR-Net
to achieve pixel-perfect segmentation (see the supplementary Figs. 1S and 2S for further details). Test results for four different architectures such as U-Net, Attention U-Net, Residual
U-Net, and proposed MADR-Net are summarized in Fig. 9. The proposed algorithm outperforms the state-of-the-art techniques with an average dice score of 89.50%, which is 3% and 2% greater
than Attention U-Net and U-Net, respectively, for skin lesion segmentation as shown in Fig. 9a. From Fig. 9b, it is observed that an average dice score of 97.31%, 97.83%, 97.78%, and 98.30%
are obtained for Attention U-Net, U-Net, Res U-Net and MADR-Net architecture for the Electron microscopy dataset. Figure 9c depicts the performance of the proposed MADR-Net architecture with
an increase of 4%, 4%, and 14% dice score with respect to Attention U-Net, U-Net, and Res U-Net. Dice scores are observed to be significantly higher for the CAMUS dataset containing LV,
Myo, and RV in Fig.9d–f. As a result, we have concluded that MADR-Net is superior to U-Net in terms of both its capacity for learning and its potential for generalization. DISCUSSION As
discussed above, to validate the performance of the proposed MADR-Net architecture, experiments were conducted for several classes of medical images which have their own set of challenges.
It must be emphasized that in all circumstances, the proposed architecture achieves convergence substantially faster. However, empirical observation revealed that after introducing the ASPP
and class-spatial attention module along with the multiscale residual block, the model was more effective in precise distinguishing of edges and texture of images, particularly in dermoscopy
and electron microscopy images. In the electron microscopy dataset, the region of interest being segmented spans the majority of the image, and thus there is a tendency to over-segment the
images. In U-Net architecture, the contextual information is passed through the skip connection which acts as a bridge between the encoder and decoder path. Further, to enhance the learning
capability, non-linear multi-scale residual blocks were used to jump over multiple layers and to restore the information loss while concatenating features from contraction and expansion
paths. To improve the segmentation accuracy over U-Net, the multi-level dilated residual block was added to the U-Net architecture to create a multi-level dilated residual U-Net. In
addition, class-spatial attention modules were also introduced to the multi-level dilated residual U-Net in order to improve the segmentation quality in terms of DSC and Jaccard index. Fig.
10 depicts the performance of the proposed architecture with three state-of-the-art techniques such as U-Net, Res U-Net, and Att U-Net on three different datasets. More specifically,
MADR-Net has a relative improvement of 5.43%, 8.73%, and 2.38% for U-Net, Attention U-Net, and Residual U-Net in terms of the Jaccard index by using the FIB-SEM dataset. MADR-Net has a
relative improvement of 3.43%, 3.92%, and 2.02% for U-Net, Attention U-Net, and Residual U-Net respectively in terms of the Jaccard index for the ISIC-2017 dataset. Similarly, for the LGG
MRI dataset, MADR-Net offered an improvement of 3.92%, 3.82%, and 12.83% Jaccard index for U-Net, Attention U-Net, and Residual U-Net, respectively. The experimental results demonstrate that
the proposed MADR-Net has significant potential to offer robust performance in evaluating single-class segmentation. For multi-class segmentation, the CAMUS dataset was used and an
improvement of 1.24%, 1.04%, and 1.66% for Epi, 0.16%, 0.32%, and 1.78% for Endo has been obtained by using U-Net, attention U-Net, and residual U-Net, respectively, in terms of dice score.
The model was trained on different loss functions, for example, dice loss, cross-entropy loss, focal Tversky loss, and a combination of these losses. From Fig. 11, we have observed that the
model achieves higher DSC for all loss functions except for the dice coefficient loss function. Furthermore, exploratory research was conducted on how the number of filters, batch size,
optimizers, and loss function impact the outcome. A remarkable improvement in segmentation was observed in both single and multi-class segmentation approaches. Results suggest the presence
of an attention mechanism, multi-scale residual block, and ASPP network improves the retention of spatial and contextual information, which is often lost during the concatenation of
features. When compared to U-Net, the proposed encoder-decoder architecture enhanced the segmentation of images. In addition to examining the best performance model for each run, we also
analyzed the model performance over each epoch. The performance analysis of the validation data on each epoch was reported in Fig. 8. Furthermore, for the four different datasets, a relative
improvement of 4.2%, 3.43%, and 3.92% was observed by using MADR-Net over U-Net. Another striking observation is that except for some minor fluctuations in the validation process, the
standard deviation of the proposed MADR-Net is substantially lower and thus demonstrates the dependability and robustness of the model. Table 5 offers a comparison of literature findings in
terms of DSC and Jaccard. It is evident that the proposed algorithm outperforms all other state-of-the-art-architectures for both single and multi-class segmentation. CONCLUSIONS In this
study, we have proposed an extended version of U-Net named multi-level attention dilated residual neural network (MADR-Net) for the segmentation of medical images. By replacing the U-Net
architecture, high-level features were extracted from multiple receptive fields and these connections improve the generalizing capability of the residual learning. The contextual features
were effectively used for the segmentation of the tumor region by leveraging the inherent property of multi-scale residual blocks and the ASPP network. On the other hand, the channel spatial
attention mechanism is of significance as it enables the retention of intricate features throughout the training process by capturing both local and global features. We have evaluated the
effectiveness of the proposed architecture on four different datasets, which further highlights the superiority of our network in comparison to baseline models. Among the handful of publicly
accessible datasets, we specifically selected those that exhibit the most unique challenges. ISIC 2017 dataset focused on skin lesion analysis which encounters challenges due to the
presence of hair strands, which makes the model difficult to delineate skin lesions and the background. The electron microscopy (FIB-SEM) dataset presents challenges for segmentation due to
the existence of artifacts and intricate biological features. There is a limited number of samples for the CAMUS dataset, which consists of three classes with 2-chamber and 4-chamber
projections of the heart. The MRI dataset consists of T1 pre-contrast, FLAIR, and T1 post-contrast MRI sequences and tumor segmentation is challenging due to the high variability of tumor
structure and location. U-Net architecture has set a benchmark for above mentioned medical image segmentation tasks. However, for complex images that are affected by artifacts and lack clear
boundaries, the MADR-Net architecture dramatically enhances performance. More specifically, from Tables 3 and 4, it is observed that the performance of MADR-Net has improved by 6%, 3%, 4%,
and 2% in comparison with U-Net architecture. Furthermore, the proposed algorithm consistently outperforms baseline architectures such as attention U-Net, and Residual U-Net for MRI, ISIC
2017, FIB-SEM, and CAMUS datasets in terms of accuracy, precision, recall, Jaccard, and dice score. The proposed MADR-Net segmentation approach not only received a higher score in the
assessment metric but was also visually more comparable to the ground truth masks. We investigated the performance of several flavors of the loss function and introduced a novel loss
function that combines cross-entropy, dice loss, and focal Tversky loss. By using this loss function, training convergence is quick and well-behaved under the presence of an imbalanced
dataset. In the future, we anticipate integrating the proposed MADR-Net model with domain-specific experts by coupling it with the post-processing stage to design better segmentation
techniques over a wide range of applications. DATA AVAILABILITY This study has been conducted using publicly available data and it can be accessed using the provided URLs: 1. CAMUS dataset:
https://www.creatis.insa-lyon.fr/Challenge/camus/, 2. ISIC dataset: https://challenge.isic-archive.com/data/, 3. Electron Microscopy dataset: https://www.epfl.ch/labs/cvlab/data/data-em/, 4.
LGG dataset: http://cancergenome.nih.gov/abouttcga/policies/informedconsent. We preprocessed the data. CODE AVAILABILITY Code of this work is available on GitHub through this link:
https://github.com/COECBT/MADR-Net. REFERENCES * Haque, I. R. I. & Neubert, J. Deep learning approaches to biomedical image segmentation. _Inform. Med. Unlocked_ 18, 100297.
https://doi.org/10.1016/j.imu.2020.100297 (2020). Article Google Scholar * Lang, R. M. _et al._ EAE/ASE recommendations for image acquisition and display using three-dimensional
echocardiography. _Euro. Heart J. Cardiovasc. Imaging_ 13(1), 1–46. https://doi.org/10.1093/ehjci/jer316 (2012). Article Google Scholar * Sharma, N. & Aggarwal, L. M. Automated medical
image segmentation techniques. _J. Med. Phys._ 35(1), 3. https://doi.org/10.4103/0971-6203.58777 (2010). Article PubMed PubMed Central Google Scholar * Xian, M. _et al._ Automatic
breast ultrasound image segmentation: A survey. _Pattern Recognit._ 79, 340–355. https://doi.org/10.1016/j.patcog.2018.02.012 (2018). Article ADS Google Scholar * Leclerc, S. _et al._
Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. _IEEE Trans. Med. Imaging_ 38(9), 2198–2210. https://doi.org/10.1109/TMI.2019.2900516 (2019). Article
PubMed Google Scholar * Lao, J. _et al._ A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. _Sci. Rep._ 7(1), 10353.
https://doi.org/10.1038/s41598-017-10649-8 (2017). Article ADS CAS PubMed PubMed Central Google Scholar * Lucchi, A., Becker, C., Márquez Neila, P., & Fua, P. Exploiting enclosing
membranes and contextual cues for mitochondria segmentation. In _Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014: 17th International Conference, Boston, MA, USA,
September 14-18, 2014, Proceedings, Part I_ _17_ 65–72 (Springer, 2014). * Gudhe, N. R. _et al._ Multi-level dilated residual network for biomedical image segmentation. _Sci. Rep._ 11(1),
14105. https://doi.org/10.1038/s41598-021-93169-w (2021). Article ADS CAS PubMed PubMed Central Google Scholar * Woo, S., Park, J., Lee, J.Y. & Kweon, I.S. Cbam: Convolutional
block attention module. In _Proceedings of the European Conference on Computer Vision (ECCV)_ 3–19 (2018) * Farahani, A. & Mohseni, H. Medical image segmentation using customized U-Net
with adaptive activation functions. _Neural Comput. Appl._ 33, 6307–6323. https://doi.org/10.1007/s00521-020-05396-3 (2021). Article Google Scholar * Lama, N., Hagerty, J., Nambisan, A.,
Stanley, R. J. & Van Stoecker, W. Skin lesion segmentation in dermoscopic images with noisy data. _J. Dig. Imaging_ https://doi.org/10.1007/s10278-023-00819-8 (2023). Article Google
Scholar * Mekuč, M. Ž, Bohak, C., Boneš, E., Hudoklin, S. & Marolt, M. Automatic segmentation and reconstruction of intracellular compartments in volumetric electron microscopy data.
_Comput. Methods Progr. Biomed._ 223, 106959. https://doi.org/10.1016/j.cmpb.2022.106959 (2022). Article Google Scholar * Meyer, C., Mallouh, V., Spehner, D., Baudrier, E., Schultz, P.
& Naegel, B. Automatic multi class organelle segmentation for cellular FIB-SEM images. In _2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI)_ 668-672 (IEEE, 2021).
https://doi.org/10.1109/ISBI48211.2021.9434075. * Shuvo, M. B., Ahommed, R., Reza, S. & Hashem, M. M. A. CNL-UNet: A novel lightweight deep learning architecture for multimodal
biomedical image segmentation with false output suppression. _Biomed. Signal Process. Control_ 70, 102959. https://doi.org/10.1016/j.bspc.2021.102959 (2021). Article Google Scholar * Long,
J., Shelhamer, E., & Darrell, T. Fully convolutional networks for semantic segmentation. In _Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition_ 3431–3440
(2015) * Ronneberger, O., Fischer, P., & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In _Medical Image Computing and Computer-Assisted Intervention–MICCAI
2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18_ 234–241. (Springer, 2015). https://doi.org/10.1007/978-3-319-24574-4_28. * Chen, L. C.,
Papandreou, G., Kokkinos, I., Murphy, K. & Yuille, A. L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. _IEEE Trans.
Pattern Anal. Mach. Intell._ 40(4), 834–848. https://doi.org/10.1109/TPAMI.2017.2699184 (2017). Article PubMed Google Scholar * Badrinarayanan, V., Kendall, A. & Cipolla, R. Segnet: A
deep convolutional encoder-decoder architecture for image segmentation. _IEEE Trans. Pattern Anal. Mach. Intell._ 39(12), 2481–2495. https://doi.org/10.1109/TPAMI.2016.2644615 (2017).
Article PubMed Google Scholar * Bermúdez-Chacón, R., Márquez-Neila, P., Salzmann, M., & Fua, P. A domain-adaptive two-stream U-Net for electron microscopy image segmentation. In _2018
IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018)_ 400–404 (IEEE, 2018). https://doi.org/10.1109/ISBI.2018.8363602. * Oktay, O., Schlemper, J., Folgoc, L.L., Lee, M.,
Heinrich, M., Misawa, K., Mori, K., McDonagh, S., Hammerla, N.Y., Kainz, B. and Glocker, B. Attention u-net: Learning where to look for the pancreas.
https://doi.org/10.48550/arXiv.1804.03999 (2018) * Ibtehaz, N. & Rahman, M. S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. _Neural
Netw._ 121, 74–87. https://doi.org/10.1016/j.neunet.2019.08.025 (2020). Article PubMed Google Scholar * Milletari, F., Navab, N. and Ahmadi, S.A. V-net: Fully convolutional neural
networks for volumetric medical image segmentation. In _2016 fourth international conference on 3D vision (3DV)_ 565-571 (IEEE, 2016). https://doi.org/10.1109/3DV.2016.79. * Alom, M.Z.,
Hasan, M., Yakopcic, C., Taha, T.M. & Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv:1802.06955.
https://doi.org/10.48550/arXiv.1802.06955 (2018) * Qin, X. _et al._ U2-Net: Going deeper with nested U-structure for salient object detection. _Pattern Recognit._ 106, 107404.
https://doi.org/10.1016/j.patcog.2020.107404 (2020). Article Google Scholar * Yan, S., Tai, X. C., Liu, J. & Huang, H. Y. Convexity shape prior for level set-based image segmentation
method. _IEEE Trans. Image Process._ 29, 7141–7152. https://doi.org/10.1109/TIP.2020.2998981 (2020). Article ADS MathSciNet Google Scholar * Abualigah, L., Almotairi, K. H. & Elaziz,
M. A. Multilevel thresholding image segmentation using meta-heuristic optimization algorithms: Comparative analysis, open challenges and new trends. _Appl. Intell._ 53(10), 11654–11704.
https://doi.org/10.1007/s10489-022-04064-4 (2023). Article Google Scholar * Van Ginneken, B., Frangi, A. F., Staal, J. J., ter Haar Romeny, B. M. & Viergever, M. A. Active shape model
segmentation with optimal features. _IEEE Trans. Med. Imaging_ 21(8), 924–933. https://doi.org/10.1109/TMI.2002.803121 (2002). Article PubMed Google Scholar * Medical and Biological Image
Analysis. BoD—Books on Demand, (2018). * Chen, X., Udupa, J. K., Bagci, U., Zhuge, Y. & Yao, J. Medical image segmentation by combining graph cuts and oriented active appearance models.
_IEEE Trans. Image Process._ 21(4), 2035–2046. https://doi.org/10.1109/TIP.2012.2186306 (2012). Article ADS MathSciNet PubMed PubMed Central Google Scholar * Arnab, A., & Torr, P.
H. Bottom-up instance segmentation using deep higher-order crfs. arXiv:1609.02583 (2016) * Georgescu, X.S., Zhou, D., Comaniciu, & Gupta, A. Database-guided segmentation of anatomical
structures with complex appearance. In _2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05)_, vol. 2, 429–436. https://doi.org/10.1109/CVPR.2005.119
(2005) * Bhandari, A. K., Singh, V. K., Kumar, A. & Singh, G. K. Cuckoo search algorithm and wind driven optimization based study of satellite image segmentation for multilevel
thresholding using Kapur’s entropy. _Exp. Syst. Appl._ 41(7), 3538–3560. https://doi.org/10.1016/j.eswa.2013.10.059 (2014). Article Google Scholar * Cheng, H. D., Jiang, X. H. & Wang,
J. Color image segmentation based on homogram thresholding and region merging. _Pattern Recogn._ 35(2), 373–393. https://doi.org/10.1016/S0031-3203(01)00054-1 (2002). Article ADS Google
Scholar * Wu, M. N., Lin, C. C., & Chang, C. C. Brain tumor detection using color-based k-means clustering segmentation. In _Third International Conference on Intelligent Information
Hiding and Multimedia Signal Processing (IIH-MSP 2007)_ vol. 2245–250. (IEEE, 2007). https://doi.org/10.1109/IIHMSP.2007.4457697 * Hesamian, M. H., Jia, W., He, X. & Kennedy, P. Deep
learning techniques for medical image segmentation: achievements and challenges. _J. Dig. imaging_ 32, 582–596. https://doi.org/10.1007/s10278-019-00227-x (2019). Article Google Scholar *
Glorot, X., & Bengio, Y. . Understanding the difficulty of training deep feedforward neural networks. In _Proceedings of the Thirteenth International Conference on Artificial
Intelligence and Statistics_ 249–256. JMLR (Workshop and Conference Proceedings, 2010). https://proceedings.mlr.press/v9/glorot10a.html. * Zhu, F., Ye, F., Fu, Y., Liu, Q. & Shen, B.
Electrocardiogram generation with a bidirectional LSTM-CNN generative adversarial network. _Sci. Rep._ 9(1), 6734. https://doi.org/10.1038/s41598-019-42516-z (2019). Article ADS CAS
PubMed PubMed Central Google Scholar * Szegedy, V., Vanhoucke, S., Ioffe, Shlens, J., & Wojna, Z., Rethinking the Inception Architecture for Computer Vision,” 2016, 2818–2826.
Accessed 18 Jun 2022; https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Szegedy_Rethinking_the_Inception_CVPR_2016_paper.html. * Pinaya, W. H. _et al._ Using deep belief
network modelling to characterize differences in brain morphometry in schizophrenia. _Sci. Rep._ 6(1), 38897. https://doi.org/10.1038/srep38897 (2016). Article ADS CAS PubMed PubMed
Central Google Scholar * de Farias, E. C. _et al._ Impact of GAN-based lesion-focused medical image super-resolution on the robustness of radiomic features. _Sci. Rep._ 11(1), 21361.
https://doi.org/10.1038/s41598-021-00898-z (2021). Article ADS CAS PubMed PubMed Central Google Scholar * Li, M., Wang, C., Zhang, H. & Yang, G. MV-RAN: Multiview recurrent
aggregation network for echocardiographic sequences segmentation and full cardiac cycle analysis. _Comput. Biol. Med._ 120, 103728. https://doi.org/10.1016/j.compbiomed.2020.103728 (2020).
Article PubMed Google Scholar * Liu, F., Wang, K., Liu, D., Yang, X. & Tian, J. Deep pyramid local attention neural network for cardiac structure segmentation in two-dimensional
echocardiography. _Med. Image Anal._ 67, 101873. https://doi.org/10.1016/j.media.2020.101873 (2021). Article PubMed Google Scholar * Ali, Y., Janabi-Sharifi, F. & Beheshti, S.
Echocardiographic image segmentation using deep Res-U network. _Biomed. Sig. Process. Control_ 64, 102248. https://doi.org/10.1016/j.bspc.2020.102248 (2021). Article Google Scholar * Guo,
L. _et al._ Dual attention enhancement feature fusion network for segmentation and quantitative analysis of paediatric echocardiography. _Med. Image Anal._ 71, 102042.
https://doi.org/10.1016/j.media.2021.102042 (2021). Article PubMed Google Scholar * Bi, L., Kim, J., Ahn, E., & Feng, D. Automatic skin lesion analysis using large-scale dermoscopy
images and deep residual networks. arXiv:1703.04197 (2017). * Zhang, G. _et al._ DSM: A deep supervised multi-scale network learning for skin cancer segmentation. _IEEE Access_ 7,
140936–140945. https://doi.org/10.1109/ACCESS.2019.2943628 (2019). Article Google Scholar * Attention-Based DenseUnet Network With Adversarial Training for Skin Lesion Segmentation. IEEE
Journals & Magazine, (IEEE Xplore, accessed 18 Jun 18 2022); https://ieeexplore.ieee.org/abstract/document/8835031. * Hasan, M. K., Dahal, L., Samarakoon, P. N., Tushar, F. I. &
Martí, R. DSNet: Automatic dermoscopic skin lesion segmentation. _Comput. Biol. Med._ 120, 103738. https://doi.org/10.1016/j.compbiomed.2020.103738 (2020). Article PubMed Google Scholar *
Tang, P. _et al._ Efficient skin lesion segmentation using separable-Unet with stochastic weight averaging. _Comput. Methods Progr. Biomed._ 178, 289–301.
https://doi.org/10.1016/j.cmpb.2019.07.005 (2019). Article Google Scholar * Zhao, X. _et al._ A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. _Med. Image
Anal._ 43, 98–111. https://doi.org/10.1016/j.media.2017.10.002 (2018). Article PubMed Google Scholar * Kamnitsas, K. _et al._ Efficient multi-scale 3D CNN with fully connected CRF for
accurate brain lesion segmentation. _Med. Image Anal._ 36, 61–78. https://doi.org/10.1016/j.media.2016.10.004 (2017). Article PubMed Google Scholar * Havaei, M. _et al._ Brain tumor
segmentation with deep neural networks. _Med. Image Anal._ 35, 18–31. https://doi.org/10.1016/j.media.2016.05.004 (2017). Article PubMed Google Scholar * Pereira, S., Pinto, A., Alves, V.
& Silva, C. A. Brain tumor segmentation using convolutional neural networks in MRI images. _IEEE Trans. Med. Imaging_ 35(5), 1240–1251. https://doi.org/10.1109/TMI.2016.2538465 (2016).
Article PubMed Google Scholar * Wadhwa, A., Bhardwaj, A. & Verma, V. S. A review on brain tumor segmentation of MRI images. _Magn. Reson. Imaging_ 61, 247–259.
https://doi.org/10.1016/j.mri.2019.05.043 (2019). Article PubMed Google Scholar * Xiao, C. _et al._ Automatic mitochondria segmentation for EM data using a 3D supervised convolutional
network. _Front. Neuroanat._ 12, 92. https://doi.org/10.3389/fnana.2018.00092 (2018). Article CAS PubMed PubMed Central Google Scholar * Mekuč, M. Ž _et al._ Automatic segmentation of
mitochondria and endolysosomes in volumetric electron microscopy data. _Comput. Biol. Med._ 119, 103693. https://doi.org/10.1016/j.compbiomed.2020.103693 (2020). Article CAS Google Scholar
* Oztel, I., Yolcu, G., Ersoy, I., White, T., & Bunyak, F. Mitochondria segmentation in electron microscopy volumes using deep convolutional neural network. In _2017 IEEE International
Conference on Bioinformatics and Biomedicine (BIBM)_ 1195–1200 (IEEE, 2017). https://doi.org/10.1109/BIBM.2017.8217827. * Zhang, Z., Liu, Q. & Wang, Y. Road extraction by deep residual
u-net. _IEEE Geosci. Remote Sens. Lett._ 15(5), 749–753. https://doi.org/10.1109/LGRS.2018.2802944 (2018). Article ADS CAS Google Scholar * Azad, R., Asadi-Aghbolaghi, M., Fathy, M.,
& Escalera, S. Bi-directional ConvLSTM U-Net with densley connected convolutions. In _Proceedings of the IEEE/CVF International Conference on Computer vision Workshops_. Accessed 21 Aug
2022; https://openaccess.thecvf.com/content_ICCVW_2019/html/VRMI/Azad_Bi-Directional_ConvLSTM_U-Net_with_Densley_Connected_Convolutions_ICCVW_2019_paper.html (2019) * He, K., Zhang, X., Ren,
S., & Sun, J. Deep residual learning for image recognition. In _Proceedings of the IEEE conference on computer vision and pattern recognition_ 770–778.
https://doi.org/10.1109/CVPR.2016.90 (2016) * Codella, N.C., Gutman, D., Celebi, M.E., Helba, B., Marchetti, M.A., Dusza, S.W., Kalloo, A., Liopyris, K., Mishra, N., Kittler, H. and Halpern,
A. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration
(isic). In _2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018)_ 168–172 (IEEE, 2018) https://doi.org/10.1109/ISBI.2018.8363547. * Lucchi, Y. Li, and P. Fua, “Learning
for Structured Prediction Using Approximate Subgradient Descent with Working Sets. In _2013 IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA_, 1987–1994.
https://doi.org/10.1109/CVPR.2013.259 (2013). * Mazurowski, M. A. _et al._ Radiogenomics of lower-grade glioma: algorithmically-assessed tumor shape is associated with tumor genomic subtypes
and patient outcomes in a multi-institutional study with The Cancer Genome Atlas data. _J. Neuro-Oncol._ 133, 27–35. https://doi.org/10.1007/s11060-017-2420-1 (2017). Article CAS Google
Scholar * Polat, H. Multi-task semantic segmentation of CT images for COVID-19 infections using DeepLabV3+ based on dilated residual network. _Phys. Eng. Sci. Med._ 45(2), 443–455.
https://doi.org/10.1007/s13246-022-01110-w (2022). Article PubMed PubMed Central Google Scholar * Zhang, Z., & Sabuncu, M. Generalized cross entropy loss for training deep neural
networks with noisy labels. Advances in neural information processing systems. Accessed 19 Jun 2022;
https://proceedings.neurips.cc/paper/2018/hash/f2925f97bc13ad2852a7a551802feea0-Abstract.html (2018) * Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S., & Jorge Cardoso, M.
Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In _Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision
Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings 3_
240–248. (Springer, 2017). https://doi.org/10.1007/978-3-319-67558-9_28. * Bertels, D., Robben, D., Vandermeulen, & Suetens, P. Optimization with soft dice can lead to a volumetric bias.
Int. MICCAI Brainlesion Workshop, 89–97, (2019). * Abraham, N., & Khan, N. M. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In _2019 IEEE
16th International Symposium on Biomedical Imaging (ISBI 2019)_ 683–687 (IEEE, 2019). https://doi.org/10.1109/ISBI.2019.8759329. * Xiao, C., Liu, J., Chen, X., Han, H., Shu, C. & Xie, Q
Deep contextual residual network for electron microscopy image segmentation in connectomics. In: _IEEE Conference Publication_, (IEEE Xplore, accessed 19 Jun 2022);
https://ieeexplore.ieee.org/abstract/document/8363597. * Goyal, M., Oakley, A., Bansal, P., Dancey, D. & Yap, M. H. Skin lesion segmentation in dermoscopic images with ensemble deep
learning methods. _IEEE Access_ 8, 4171–4181. https://doi.org/10.1109/ACCESS.2019.2960504 (2019). Article Google Scholar * Su, R., Zhang, D., Liu, J. & Cheng, C. MSU-Net: Multi-scale
U-Net for 2D medical image segmentation. _Front. Genet._ 12, 639930. https://doi.org/10.3389/fgene.2021.639930 (2021). Article PubMed PubMed Central Google Scholar * Naser, M. A. &
Deen, M. J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. _Comput. Biol. Med._ 121, 103758. https://doi.org/10.1016/j.compbiomed.2020.103758
(2020). Article PubMed Google Scholar * Chao, C. J. _et al._ Comparative eminence: Foundation versus domain-specific model for cardiac ultrasound segmentation. _medRxiv_
https://doi.org/10.1101/2023.09.19.23295772 (2023). Article PubMed PubMed Central Google Scholar * Leclerc, S., Smistad, E., Grenier, T., Lartizien, C., Ostvik, A., Cervenansky, F.,
Espinosa, F., Espeland, T., Berg, E.A.R., Jodoin, P.M. & Lovstakken, L. RU-Net: a refining segmentation network for 2D echocardiography. In _2019 IEEE International Ultrasonics Symposium
(IUS)_ 1160–1163 (IEEE, 2019). https://doi.org/10.1109/ULTSYM.2019.8926158. * Upadhyay, S., Beevi, A.S. & Kalady, S. Left Ventricle segmentation of 2D Echocardiography using deep
learning. In _International Conference on Computer Vision and Image Processing_ 87–98 (Springer, 2022). https://doi.org/10.1007/978-3-031-31407-0_7. * Sfakianakis, C., Simantiris, G. &
Tziritas, G. GUDU: Geometrically-constrained Ultrasound Data augmentation in U-Net for echocardiography semantic segmentation. _Biomed. Signal Process. Control_ 82, 104557.
https://doi.org/10.1016/j.bspc.2022.104557 (2023). Article Google Scholar Download references FUNDING This work was funded by multiple grants from the Department of Biotechnology, Ministry
of Science and Technology (BT/COE/34/ SP15097/2015, PI: Anurag S. Rathore; BT/PR34224/Al/133/10/2O19, PIs: Anurag S. Rathore and Sandeep Seth) and Partnership 2020 Grant (PIs: Rohit
Bhargava and Anurag S. Rathore). This study was supported by the Tower project (RP04636N, PI: ASR). This study used the high-performance computing capabilities of IIT Delhi. The authors
would like to express their gratitude to the Yardi School of Artificial Intelligence for their valuable assistance with this project. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Yardi
School of Artificial Intelligence, Indian Institute of Technology Delhi, Hauz Khas, New Delhi, 110016, India Keerthiveena Balraj, Manojkumar Ramteke & Anurag S. Rathore * Department of
Chemical Engineering, Indian Institute of Technology Delhi, Hauz Khas, New Delhi, India Manojkumar Ramteke & Anurag S. Rathore * Department of Laboratory Medicine and Pathology, School
of Medicine, University of Washington, Seattle, WA, USA Shachi Mittal * Departments of Bioengineering, Electrical and Computer Engineering, Mechanical Science and Engineering, Chemical and
Biomolecular Engineering and Chemistry, Beckman Institute for Advanced Science and Technology, Cancer Center at Illinois, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA
Rohit Bhargava Authors * Keerthiveena Balraj View author publications You can also search for this author inPubMed Google Scholar * Manojkumar Ramteke View author publications You can also
search for this author inPubMed Google Scholar * Shachi Mittal View author publications You can also search for this author inPubMed Google Scholar * Rohit Bhargava View author publications
You can also search for this author inPubMed Google Scholar * Anurag S. Rathore View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS BKV: Data
Curation, Formal Analysis, Software, Writing—Original Draft. MR: Conceptualization, Methodology, Investigation, Writing—Review and Editing. SM: Investigation, Formal Analysis, Validation.
RB: Investigation, Formal analysis, Validation. ASR: Conceptualization, Methodology, Investigation, Writing—Review and Editing CORRESPONDING AUTHOR Correspondence to Anurag S. Rathore.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional
claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative
Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the
original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in
the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your
intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence,
visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Balraj, K., Ramteke, M., Mittal, S. _et al._ MADR-Net: multi-level attention
dilated residual neural network for segmentation of medical images. _Sci Rep_ 14, 12699 (2024). https://doi.org/10.1038/s41598-024-63538-2 Download citation * Received: 03 October 2023 *
Accepted: 29 May 2024 * Published: 03 June 2024 * DOI: https://doi.org/10.1038/s41598-024-63538-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
KEYWORDS * Deep learning * Semantic segmentation * Convolutional neural networks * Class-spatial attention module * Atrous convolution