Establishment and validation of a redox-related long non-coding rnas prognostic signature in head and neck squamous cell carcinoma
Establishment and validation of a redox-related long non-coding rnas prognostic signature in head and neck squamous cell carcinoma"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Reduction and oxidation (redox) reactions occur in living organisms as part of normal cellular metabolism. Here, we established a novel redox-related long non-coding RNAs
(rrlncRNAs) signature to predict the prognosis and therapeutic response in Head and neck squamous cell carcinoma (HNSCC). The expression profile and clinical information were obtained from
the TCGA project. In total, 10 differently expressed rrlncRNAs associated with prognosis were identified and involved in a prognostic risk score signature by the least absolute shrinkage and
selection operator penalized Cox analysis. The area under the receiver operating characteristic curves of the survival rates predicted by the rrlncRNAs signature over one, two, and three
years were found to be 0.651, 0.670, and 0.679. Following the completion of the Kaplan–Meier survival study, we discovered that the lower-risk cohort exhibited a much longer overall survival
period in contrast with the higher-risk cohort. Univariate and multivariable Cox regression analyses demonstrated that the risk score independently served as a significant predictive
factor. GO annotation and KEGG pathway analyses illustrated that the rrlncRNAs signature was strongly associated with immune-related functions as well as signaling pathways. The
tumor-infiltrating immune cells, tumor microenvironment, immune-related functions, HLA gene family expression, immune checkpoint genes expression, and somatic variants differed substantially
between the low- and high-risk cohorts. Moreover, patients in low-risk group were predicted to present a favorable immunotherapy responsiveness, while in contrast, the high-risk group
patients might have a stronger sensitivity to “docetaxel”. According to our findings, the rrlncRNAs signature showed an excellent prognosis predictive value and might indicate therapeutic
response to immunotherapy in HNSCC. SIMILAR CONTENT BEING VIEWED BY OTHERS NON-CODING RNA-RELATED FCGBP DOWNREGULATION IN HEAD AND NECK SQUAMOUS CELL CARCINOMA: A NOVEL BIOMARKER FOR
PREDICTING PACLITAXEL RESISTANCE AND IMMUNOSUPPRESSIVE MICROENVIRONMENT Article Open access 23 February 2024 IDENTIFYING PROGNOSTIC BIOMARKERS IN ORAL SQUAMOUS CELL CARCINOMA: AN INTEGRATED
SINGLE-CELL AND BULK RNA SEQUENCING STUDY ON MITOPHAGY-RELATED GENES Article Open access 28 August 2024 MIR-378A-5P EXERTS TUMOR-SUPPRESSIVE EFFECTS ON ESOPHAGEAL SQUAMOUS CELL CARCINOMA
AFTER NEOADJUVANT IMMUNOTHERAPY BY DOWNREGULATING APOC1/CEP55 Article Open access 03 January 2024 INTRODUCTION Head and neck squamous cell carcinoma (HNSCC) is ranked as the 6th most
prevalent malignant tumor on a global scale and the incidence is still continuously increasing1,2. Risk factors of the development of HNSCCs include infection with human papillomavirus
(HPV), exposure to environmental pollutants, alcohol abuse, and tobacco smoking3,4. At present, the most commonly utilized therapeutic approaches for HNSCC are chemotherapy, radiotherapy,
and surgery. In addition, immunotherapy, as an emerging strategy for cancer treatment, has also been well represented in the treatment of HNSCC5,6,7. According to studies, pembrolizumab
monotherapy is a suitable first-line treatment for patients with PD-L1-positive recurrent or metastatic HNSCC and pembrolizumab combined with chemotherapy is a suitable first-line treatment
for recurrent or metastatic HNSCC patients8. Nivolumab therapy resulted in prolonged overall survival among patients with platinum-refractory, recurrent HNSCC9. In the last 3 decades, the
rate of survival for HNSCC patients has increased to a certain extent as a result of advancements in medicine along with breakthroughs in molecular biology10. However, due to the
asymptomatic nature of early lesions and the risk of recurrence and metastasis, the five-year rate of survival of HNSCC patients is still low at an average of less than 50%11,12. Thus, it is
of significant clinical importance to develop biomarkers related to the prognosis of HNSCC. Reduction and oxidation (redox) reactions occur in living organisms as part of normal cellular
metabolism, reactive oxygen species (ROS) are generated by these redox events physiologically or pathologically13. A redundancy of ROS can induce oxidative damage of macromolecules including
proteins, lipids, and DNA, then further enhance the incidence as well as the development of tumors in multistage14. Over the past few years, research reports have explored the function
played by redox-related products in different cancers and demonstrated the promising value for predicting the prognosis of patients. It was observed that in advanced cervical cancer, the
antioxidant levels were significantly declined, while the levels of lipid peroxidation were remarkably increased compared to healthy controls15. The researchers also suggested the evaluation
of oxidant and antioxidant levels could help to anticipate the treatment responsiveness of advanced cervix cancer to neoadjuvant chemoradiation16. By measuring the levels of superoxide
dismutase (SOD) activity, nitric oxide (NO), glutathione (GSH), and lipid peroxidation products (LPO) in blood, we discovered that there was an increase in oxidative stress and a
simultaneous decrease in antioxidant enzyme activity with the advance in the lung cancer stages17. Xiang et.al. demonstrated that selenite induces cell death and apoptosis by the production
of superoxide and altering intracellular redox state in human prostate cancer cells18. Long non-coding RNAs (lncRNAs) are a subset of RNAs with a length exceeding 200 nucleotides, often
overlapping with, or interspersed between multiple coding and non-coding transcripts19. It has been revealed that lncRNAs were involved in modulating gene expression via transcription
processing, post-transcriptional processing, as well as chromatin alteration20. Previous researchers have established some lncRNA signatures, which have excellent prognostic prediction
values in various cancers21. The reported signatures included lncRNAs with specific functions, such as ferroptosis-related lncRNAs, autophagy-related lncRNAs, some differentially expressed
lncRNAs, and immune-related lncRNAs22. Lately, a promising signature comprised 9 redox-related lncRNAs in renal clear cell carcinoma was reported, which had the area under ROC curves (AUCs)
demonstrating the prediction capacity for survival rates over one, three, and five years whose values were found to be 0.780, 0.746, and 0.76423. However, there are no redox-related lncRNAs
signatures that have been reported for prognostic prediction in HNSCC so far. In the present research, we established an innovative ten-redox-related lncRNAs prognostic signature in HNSCC.
Furthermore, we investigated the relationship between the signature and clinicopathological characteristics, tumor immune landscape, and somatic variants. Moreover, the predictive values in
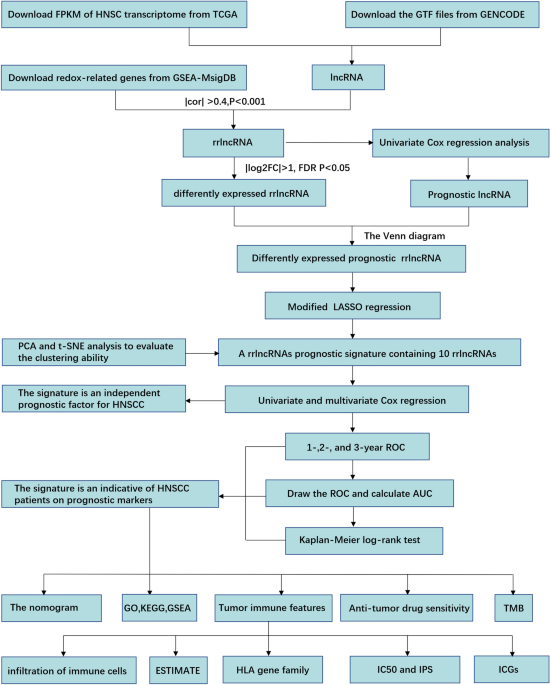
prognosis and response to immunotherapy and chemotherapy were assessed. MATERIALS AND METHODS DATA RESOURCE The Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov) was
utilized to acquire gene transcript data from high-throughput sequencing (HTSeq) with normalization in Fragments Per Kilobase of transcript per Million mapped reads (FPKM) as well as simple
nucleotide variation data of TCGA-HNSC project. Moreover, clinicopathological and survival data were obtained. By utilizing the GENCODE human website (https://www.gencodegenes.org/human/),
we acquired the GTF file of lncRNA annotation (version GRCh38.p13) used in the present research. The redox-related genes (rrGenes) were retrieved from the website of GSEA-MSigDB
(https://www.gsea-msigdb.org/gsea/msigdb) according to “redox” as the keyword. The Pearson correlation coefficient was utilized to identify the redox-related lncRNAs having correlation
coefficients more than 0.4 and P less than 0.001. Then, we determined that |log2FC|> 1 and false discovery rate (FDR) _P _< 0.05 were the significant criteria for the definition of
differently expressed lncRNAs. By means of univariate Cox regression analysis, we determined the prognostic significance of lncRNAs (_P _< 0.05). The Venn diagram was performed to overlap
the lncRNAs in the differently expressed cluster and prognostic cluster. Given the fact that no personally identifiable information was utilized in the present research, the Institutional
Review Board of the Ningbo Women and Children’s Hospital approved the exemption from the need for ethical clearance for the present research. The study flow was showed in Fig. 1.
ESTABLISHMENT OF A REDOX-RELATED LNCRNAS PROGNOSTIC SIGNATURE The least absolute shrinkage and selection operator (LASSO) penalized Cox regression analyses were utilized to verify and build
a redox-related lncRNAs prognostic signature. The risk score of each patient was computed utilizing the equation as follows: Risk score = Coefficient lncRNA1 × Expression lncRNA1 +
Coefficient lncRNA2 × Expression lncRNA2 + Coefficient lncRNA3 × Expression lncRNA3 + …… + Coefficient lncRNAn × Expression lncRNAn. The patients were divided into low- and high-risk cohorts
according to their median risk value. The t-SNE and PCA algorithms were employed to evaluate the clustering ability of this risk score signature. To anticipate the prognosis of HNSCC
patients, the one-, three-, and five-year ROC curves of signature were constructed, and the results were compared with standard clinicopathological criteria. Subsequently, we utilized the
decision-curve analysis (DCA) plot to assess the normalized overall advantage of the signature and performed a comparison between its benefits and the benefits of other clinical parameters.
The Kaplan–Meier method was utilized to create survival curves for various risk cohorts, followed by a comparison. A further investigation was carried out to determine if the rrlncRNAs
signature independently served as a prognostic indicator in HNSCC patients utilizing univariate and multivariable Cox regression models. Kaplan–Meier survival curves of subcohort analyses
according to age (< 60, ≥ 60), gender (female, male), T stage (T1-2, T3-4), histologic grade (G1-2, G3), lymphatic metastasis (N0, N1-3), and clinical stage (stage I-II, stage III-IV)
were severally generated. CORRELATION ANALYSIS OF THE RRLNCRNAS SIGNATURE WITH CLINICAL VARIABLES AND ESTABLISHMENT OF A NOMOGRAM In the present research, we utilized the chi-squared test to
delve into the relationship between the clinicopathological characteristics and risk score, which was represented as a heatmap. The _rms_ package of the R software was utilized to construct
the nomogram that integrated the risk score and clinical and pathological features for evaluating the HNSCC patient's survival rates over one, three, and five years
(https://CRAN.R-project.org/package=rms). ROC curves of the nomogram were conducted to evaluate the predictive ability in anticipating the OS rates over one, three, and five years, and the
calibration chart was drawn to examine the accuracy of the nomogram. FUNCTION ENRICHMENT ANALYSIS Enrichment analyses on the basis of differentially expressed genes (DEGs) between low and
high-risk cohorts were conducted to explore the underlying functional pathways. The threshold value of significantly differential expression was set at FDR _P _< 0.05 and |log2FC|≥ 1.
Gene ontology (GO) function analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed based on DEGs24,25. Gene set enrichment analysis (GSEA, version 4.1.0) based
on the annotated gene set file (c2.cp.kegg.v7.0.symbols.gmt) was executed to investigate the potential signaling pathways between low and high-risk cohorts in HNSCC. TUMOR IMMUNE FEATURE
Several acknowledged algorithms, such as EPIC, MCPcounter, quanTIseq, CIBERSORT-ABS, xCELL, CIBERSORT, and TIMER were employed to evaluate the discrepancies of tumor-infiltrating immune
cells (TIICs) between high- and low-risk cohorts. ESTIMATE algorithm is a method for obtaining ESTIMATE, stromal, and immune scores based on the expression of related molecular biomarkers in
immune and stromal cells, which was applied for assessing the tumor microenvironment (TME) disparities in high and low-risk cohorts26. Furthermore, single-sample gene set enrichment
analysis (ssGSEA) was further used for exploring the immune-related function differences between low and high-risk cohorts. Moreover, the expression divergences of the human leukocyte
antigen (HLA) gene family and common immune checkpoint genes (ICGs) retrieved from previous literature27 in different risk cohorts were also assessed_._ RESPONSE PREDICTION TO CHEMOTHERAPY
AND IMMUNOTHERAPY To examine the clinical application prospect of the rrlncRNAs signature, we investigated the prediction power of patient prognosis in responding to chemotherapy and
immunotherapy in HNSCC. The 50% inhibiting concentration (IC50) values of 4 conventional chemotherapeutic drugs, including “docetaxel”, “cisplatin”, “gemcitabine”, and “pacitaxel” between
high and low-risk cohorts, were inferred by utilizing the pRRophetic algorithm. Besides, immunophenoscore (IPS) was calculated to anticipate the potential clinical immunotherapy
responsiveness in patients with HNSCC. IPS analysis was a bioinformatics method to quantitatively score the tumor immunogenicity range from 0 to 10, which has been verified for predicting
the clinical response to treatment of immune checkpoint inhibitors (ICIs)28. In principle, a lower IC50 value and higher IPS value portend a better sensitivity to chemotherapy and
immunotherapy, respectively. ASSOCIATION BETWEEN THE SOMATIC VARIANTS AND RISK SCORE SIGNATURE We identified the topmost twenty commonly mutated genes of HNSCC samples, followed by mutation
frequency comparison between high and low-risk cohorts. To calculate tumor mutation burden (TMB), we examined the quantity of somatic, coding, indel, and base substitutions per megabase (Mb)
of the genome for subsequent analyses. The TMB score of each sample was calculated by dividing the overall number of mutations counted by the size of the exome used in the calculation
process. The size of the exome was estimated to be 38 megabases (Mb)29. Furthermore, the association between TMB levels and OS time in HNSCC patients was examined in the present research.
Moreover, the prognostic significance of the rrlncRNAs signature was assessed in the low and high TMB subcohorts. STATISTICAL ANALYSIS The GESA and HTSeq FPKM data preparation materials were
extracted and structured utilizing the Perl software (version: 5.32). The analyses were carried out utilizing the R software (R: A language and environment for statistical computing. R
Foundation for Statistical Computing, Vienna, Austria; version: 4.0.3, https://www.R-project.org/) with specialized packages. Statistical significance was determined when the _p-_value was
less than 0.05. ETHICS STATEMENT TCGA is a publicly accessible database. Ethical permission has been acquired for the patients in the databases. Users may get relevant information for free
and use it to conduct research as well as publish pertinent publications. Due to the fact that the present research used open-source data, there are no ethical concerns or potentially
conflicting interests. The Lihuili Hospital's Institutional Review Board granted its exemption from ethical clearance. RESULTS DATA CHARACTERISTICS The expression profile of 499 HNSCC
tissues as well as 44 adjoining normal tissues was included in the present research. Table 1 depicts the patients’ clinical variables, such as gender, age, histologic grade, T stage,
lymphatic metastasis, clinical stage, and survival status. In total, 55 redox-related genes (rrGenes) were downloaded and included in the analysis (Table S1). In all, 636 lncRNAs were
identified as redox-related lncRNAs (rrlncRNAs) (Table S2). Following with identification of 172 differentially expressed rrlncRNAs and 108 prognostic rrlncRNAs, 31 overlapped rrlncRNAs in
the two clusters were extracted, which were used for further analyses (Fig. 2A, B) (Table S3). CONSTRUCTION OF A RRLNCRNAS RISK SCORE PROGNOSTIC SIGNATURE According to the least absolute
shrinkage and selection operator (LASSO) penalized Cox analysis, 10 out of 31 rrlncRNAs (AC133644.1, MIR4435-2HG, PTOV1-AS2, AL355488.1, LINC02446, DBH-AS1, AC010226.1, Z97653.1, LINC02084,
AC098487.1) were identified and used to construct a risk score prognostic signature (Table S4). After that, the relationship network between identified rrlncRNAs and relevantly co-expressive
rrGenes (_PRDX5_, _EGLN2_, _TXN2_, _PRDX4_, _CYBA_, _GPX1_, _VASN_, _HVCN1_, _NCF4_, _ARHGDIB_) was constructed (Fig. 2C). The results of PCA (Fig. 3A) and t-SNE (Fig. 3B) analyses showed a
favorable clustering power of this rrlncRNAs prognostic signature in HNSCC. Using the risk assessment model for survival status predictions, it was discovered that an increment in the risk
score was associated with a substantial rise in the number of patients who died (Fig. 3C). The area under the ROC curves (AUCs) of the risk score signature for survival rates over one, two,
and three years were found to be 0.651, 0.670, and 0.679 (Fig. 3D). The AUC value of our signature was positively superior to age (0.580), gender (0.500), grade (0.544), and stage (0.559) in
predicting the survival of HNSCC (Fig. 3E). The result of a decision-curve analysis (DCA) plot illustrated that the risk score signature had a most remarkable standardized net benefit
compared to other clinical factors, such as stage, gender, grade, and age (Fig. 3F). According to the findings from the Kaplan–Meier survival analysis, the low-risk cohort exhibited a
considerably longer overall survival time as opposed to the high-risk cohort (_P _< 0.001, Fig. 3G). These findings revealed that the established rrlncRNAs risk score prognostic signature
had a promising clinical prediction efficiency in patients with HNSCC. Moreover, the rrlncRNAs risk score signature was confirmed to independently function as a prognostic marker with the
aid of univariate (Fig. 3H) and multivariate Cox regression analyses (Fig. 3I). CORRELATIONS BETWEEN THE RISK SCORE SIGNATURE AND CLINICAL CHARACTERISTICS The subgroup analyses of the
prognostic prediction effect illustrated that the low-risk cohort had longer OS time as opposed to the high-risk cohort in all subgroups except for the female group (Fig. 4A–F). The
correlation analysis revealed that stage III/IV and T3/4 patients exhibited an elevated risk score in contrast with the stage I/II and T1/2 patients (Fig. 4G). Furthermore, a nomogram that
incorporated the risk score together with common prognostic clinicopathological factors (gender, N stage, T stage, histologic grade, age, and clinical stage) was successfully generated for
estimating the 1-, 3-, 5-year survival probability (Fig. 5A). The AUC of the nomogram for anticipating the OS rates over one, three, and five years were found to be 0.683, 0.756, and 0.742
(Fig. 5B). The calibration plots for OS over one, three, and five years indicated the nomogram showed a promising predictive capacity and performance for HNSCC (Fig. 5C). POSSIBLE MECHANISM
THAT UNDERLIES THE EFFECTS OF PROGNOSTIC RRLNCRNAS SIGNATURE The results of GO and KEGG pathway analyses illustrated that the rrlncRNAs signature was strongly correlated with immune-related
functions and signaling pathways. The immune-related signaling pathways in biological processes (BP) mainly included “immune response-activating cell surface receptor signaling pathway”,
“immune response-activating signal transduction”, “humoral immune response”, “adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin
superfamily domains”, “lymphocyte-mediated immunity”, “B cell-mediated immunity”, “immunoglobulin mediated immune response”, “humoral immune response mediated by circulating immunoglobulin”;
in molecular functions (MF) included “immunoglobulin complex”, “immunoglobulin complex, circulating”, “T cell receptor complex”, “immunological synapse”; in cellular components (CC)
included “antigen-binding”, “immunoglobulin receptor binding”, “immune receptor activity” (Fig. 6A). KEGG pathway analyses revealed additional immune-related signaling pathways, such as
“primary immunodeficiency”, “T cell receptor signaling pathway”, “Th17 cell differentiation”, “Th1 and Th2 cell differentiation”, “PD-L1 expression and PD-1 checkpoint pathway in cancer”,
“Natural killer cell mediated cytotoxicity”, and so on (Fig. 6B). The findings from GSEA illustrated that the high-risk score cohort exhibited a significant inverse association with several
immune-related signaling pathways, which included “antigen processing and presentation”, “B cell receptor signaling pathway”, “FC epsilon RI signaling pathway”, “FC gamma R mediated
phagocytosis”, “intestinal immune network for IGA production”, “natural killer cell-mediated cytotoxicity”, “primary immunodeficiency”, and “T cell receptor signaling pathway” (Fig. 6C).
ASSOCIATION BETWEEN THE TUMOR IMMUNE LANDSCAPE AND RRLNCRNAS SIGNATURE The findings illustrated that the low-risk cohort had a greater proportion of activated immune cells infiltrating which
included macrophage, CD4 + T cells, neutrophil, CD8 + T cells, myeloid dendritic cell, B cell, and activated NK cell (Fig. 7A). Furthermore, the ESTIMATE algorithm revealed that the
low-risk score cohort exhibited elevated immune and ESTIMATE scores, indicating a higher infiltration of immune cells and attenuated tumor cell purity in the low-risk cohort (Fig. 7B).
Immune-related function analyses showed that checkpoint, cytolytic (activity), inflammation-promoting, MHC class I, HLA, T cell co-stimulation, APC co-stimulation, T cell co-inhibition, CCR,
APC co-inhibition, and type II INF response were significantly activated in the low-risk cohort (Fig. 7C). Additionally, the expression of the human leukocyte antigen (HLA) gene family was
observably elevated in the low-risk score cohort as opposed to the high-risk score cohort, except for _HLA-A_, _HLA-G_, _HLA-J_, _HLA-L_, and _HLA-H_ (Fig. 7D). The low-risk cohort also had
substantially upmodulated immune checkpoint genes expression, which included _CD8A_, _GZMA_, _PRF1_, _HAVCR2_, _IFNG_, _GAMB_, _CD274_, _TNF_, _IDO1_, _LAG3_, _CTLA4_, and _PDCD1_ (Fig. 7E).
PREDICTIVE EFFECTS OF THE RRLNCRNAS SIGNATURE FOR THERAPEUTIC SENSITIVITY The predictive effect of the rrlncRNAs signature in chemotherapy was evaluated according to the 50% inhibiting
concentration (IC50) values of 4 conventional chemotherapeutic drugs, including “docetaxel”, “cisplatin”, “gemcitabine”, and “pacitaxel”. The findings illustrated that the IC50 value of
“docetaxel” was considerably elevated in the low-risk cohort as opposed to the high-risk cohort, but there were no substantial differences among the other three drugs. This suggests that the
high-risk cohort patients might have a stronger sensitivity to “docetaxel” (Fig. 8A). Besides, the prediction capacity of the rrlncRNAs signature in immune checkpoint inhibitors (ICIs) was
evaluated by immunophenoscore (IPS) algorithm. The results demonstrated that the IPS value in the low-risk cohort was considerably elevated in contrast with the high-risk cohort in various
ICIs conditions, including “CTLA4_pos_PD1_pos”, “CTLA4_pos_PD1_neg”, and “CTLA4_neg_PD1_pos”. These results suggest a probably favorable immunotherapy responsiveness in the low-risk cohort
(Fig. 8B). DISCREPANCY OF SOMATIC VARIANTS IN DIFFERENT RISK SCORE COHORT We explored the topmost twenty commonly mutated genes in HNSCC, and the five of the most commonly mutated genes
included _FAT1_, _TTN_, _CDKN2A_, _TP53_, and _MUC16_. Furthermore, disparities in mutation rates of these genes were evaluated in low- and high-risk cohorts. These findings demonstrated
that _NOTCH1_ and _TP53_ mutations were highly frequent in the high-risk cohort whereas _CSMD3_, _PIK3CA_, _NSD1_, _USH2A_ had obviously increased frequent mutations in the low-risk cohort
(Fig. 9A). Moreover, patients exhibiting attenuated TMB levels had significantly longer survival times as opposed to the ones having high TMB (Fig. 9B). We examined the prognostic value of
the rrlncRNAs signature in high and low-TMB cohorts. Finally, we discovered that the prognostic signature had similar strong prognostic significance in both the high- and low-TMB cohorts,
demonstrating that the TMB status had no influence on the prediction (Fig. 9C). DISCUSSION HNSCC is among the most prevalent contributor to cancer mortality globally. Due to the lack of
early screening and the common invasion of regional lymph nodes, most cases are detected at advanced stages, with an unfavorable 5-year survival rate1,30. Although HPV detection became an
important indicator to guide treatment and predict prognosis in HNSCC31, however, due to the tumor heterogeneity, more biomarkers are expected to provide better prognostic and therapeutic
efficacy prediction. In HNSCC, accumulation or suppression of ROS occurred at all stages of disease underlying tumorigenesis, development, progression, and response to various therapies on
account of alterations in redox metabolism32. For the last few years, studies have shown that modulators of redox metabolism were promising strategies for clinical management of HNSCC,
including detection and diagnosis, synergizing the efficacy of conventional radiotherapy, chemotherapy, EGFR targeted therapy, and immunotherapy32,33. The novel role of specifical lncRNAs
signatures as prognosis predictors have been reported in a variety of cancers including breast cancer34, laryngeal cancer35, lung cancer36, colorectal cancer37, and HNSCC38. Here, we
established a redox-related lncRNAs signature as a prognostic biomarker for HNSCC patients, which comprised ten lncRNAs related to redox genes, including AC133644.1, PTOV1-AS2, AL355488.1,
LINC02446, DBH-AS1, AC010226.1, Z97653.1, LINC02084, MIR4435-2HG, and AC098487. Research reports have demonstrated the role of lncRNA MIR4435-2HG as an oncogenic lncRNA in various cancers.
Dong et al. found an elevated expression of MIR4435-2HG in colorectal cancer (CRC) tissue and promoted CRC growth as well as metastasis via miR-206/YAP1 axis39. Gao et al. discovered that
MIR4435-2HG is enhanced in gastric cancer cells and that it promotes the proliferation as well as invasiveness of these cells via targeting the miR-138-5p/Sox4 axis40. MIR4435-2HG was
observed to be markedly upregulated in HNSCC tissues, which could promote cancer progression by modulating the miR-383-5p/RBM3 axis41. DBH-AS1 was discovered to be a risk lncRNA associated
with an unfavorable prognosis in hepatocellular carcinoma42. The rrlncRNA signature showed a promising clinical prediction efficiency and could serve as a prognostic factor in HNSCC.
However, the functions of the other eight lncRNAs involved in the signature deserve further exploration. To investigate the potential biological functions as well as signaling pathways by
which our rrlncRNAs signature works in carcinogenicity, GO and KEGG enrichment analyses were performed according to DEGs between the low and high-risk cohorts. The findings demonstrated that
the rrlncRNAs signature was substantially enriched in many immune and immune response-related biological functions and signaling pathways. Actually, a recent review summarized the effects
of redox signaling pathways on the immune cells' function regulation in cancer43. Evidence revealed that ROS and other main cellular antioxidant pathways were closely related to the
proliferation, survival, and regulation of the function of T cells, B cells, and macrophages. A review pinpointed operational concepts related to the redox biology network applied to the
pathophysiology and therapeutics of solid tumors, including radiotherapy, photodynamic therapy, and anti-tumor immune therapy. The authors draw attention on the consequences of the damage
signals delivered by oxidative stress-injured cells to neighboring and distant cells, and emphasize the benefits of therapeutically triggered immunologic cell death in metastatic cancer44.
David D Roberts reported the role of cellular redox signaling in T cell proliferation and activation45. Roberta Vené revealed the vital function of redox remodeling in B-cell activation and
differentiation46. Xiang Li found that ROS decreased the amount of tumor exo-miR-155 that was taken up by macrophages, resulting in enhanced macrophage infiltration and T cell inactivation
characterized by upregulation of PD-L1 in ovarian cancer47. According to these results, we speculated that the rrlncRNAs signature might have a promising impact on the tumor immune milieu in
HNSCC. Therefore, we further investigated the associations between the established signature and tumor immune landscape in HNSCC. TIICs, a significant constituent of the tumor
microenvironment (TME), is shown to perform an integral function in cancer prognosis and response to immune therapy in a variety of tumor types, including lung cancer48,49, breast cancer50,
colorectal cancer51 and so on. For patients with HNSCC, research conducted by Zhou et.al. suggested the cell structure of TIICs is remarkably different in HNSCC with or without HPV infection
and the variances are crucial for the therapeutic outcome and the development of the tumor52. In the present research, our analysis of immune-infiltrated cells enrichment showed a
significant discrepancy between low- and high-risk score cohorts, suggesting that the prognosis predictive effect may be correlated with its clustering efficiency in tumor immune features.
TME score evaluated by ESTIMATE analysis demonstrated that the low-risk score cohort exhibited elevated immune and ESTIMATE scores, indicating a greater tumor purity in the high-risk cohort.
Immune-related function analyses showed that checkpoint, cytolytic (activity), T cell co-inhibition, APC co-inhibition, T cell co-stimulation, inflammation-promoting, HLA, MHC class I, APC
co-stimulation, CCR, and type II INF response differed substantially between the high- and low-risk score cohorts.HLA class I might be additionally subdivided into HLA-A, -B, and -C
subclasses based on the location of the encoding gene in each of these subclasses. HLA class I molecules are essential for presenting tumor antigens to naïve T cells, whereas HLA class II
molecules stimulate the conversion of these naïve T cells into activated T cells by delivering exogenous antigen peptides to CD4 + T cells53,54. Actually, according to the findings of
numerous research reports, the expression of MHC on tumor cells serves as an effective surrogate indicator for the general immunogenicity of the tumor and also a predictive indicator for
immunotherapy response55. As shown in the results of two distinct clinical trials, CheckMate 064 and CheckMate 069, attenuated expression of tumor HLA class I molecule (≤ 30%) was linked to
an absence of response to ipilimumab, while increased expression of tumor HLA class II molecule (> 1 percent) was associated with greater tumor responsiveness to nivolumab56. In the
present research, we discovered that the expression of the human leukocyte antigen (HLA) gene family was observably elevated in the low-risk score cohort as opposed to the high-risk score
cohort, suggesting the predictive value of the signature in response to immune therapy. Notablely, we found that the patients in high-risk cohort might have a stronger sensitivity to
“docetaxel”. Similar studies have shown that the sensibility to “docetaxel” was higher in the high-risk patients in HNSCC based on a hypoxia-related lncRNA model and a cuproptosis-related
lncRNA signature57,58. Wei Zhang et al. revealed that lncRNA LINC00184 could promote “docetaxel” resistance and immune escape in prostate cancer cells and lead to an increase in PD-L1
expression59. We speculate that the lncRNAs involved in our signature might influence the sensitivity of cancer cells to docetaxel by regulating redox reaction, hypoxia, cuproptosis, immune
microenvironment and other factors, which is worth further exploration and research.At present, the main mechanism of immunotherapy is focused on the blocking of immune checkpoints. In the
present research, we discovered that HNSCC with a low-risk score exhibited significantly upmodulated expression of immune checkpoint genes including _CD8A_, _HAVCR2_, _LAG3_, _IFNG_
(_IFNγ_), _GZMB_, _CTLA4_, _TNF_, _CD274_, _PRF1_, _GZMA_, and _PDCD1_. _PDCD1_ (_PD1_), _CD274_ (_PD-L1_), and _CTLA4_ are three already known immune checkpoints, numerous studies have
achieved great success in immunotherapy targeting _PD1_/_PDL1_ therapy in different solid tumors in the past decade60, and other immune checkpoint inhibitors have also made substantial
progress. _IDO1_ is a new immunological checkpoint target that is defined as a rate-limiting metabolic enzyme responsible for transforming tryptophan into kynurenines. Despite the growing
evidence that blocking _IDO1_ alone does not produce therapeutic benefit, there is convincing evidence for the use of combined methods to deliver a synergistic advantage to patients61. One
recent study revealed that as _IDO1_ expression level increases, so do other immune checkpoints, including _PD-L2_, _CTLA-4_, _PD-L1_, _PD-1_, and so on. In addition, one research showed the
high expression of _IDO1_ was induced by IFNγ that inhibited the STAT1 signaling pathway cooperatively, which led to suppressing the process of cell death and activating the dormancy
program62. _GZMA_ and _PRF1_ are two key cytolytic effectors, tightly co-expressed, dramatically upregulating upon CD8 + T cell activation. It has been demonstrated that the geometric mean
of _PRF1_ and _GZMA_ expression is correlated with both the neoantigen load and the local immune infiltrates cytolytic activity63. _HAVCR2_ also named _TIM3_, was expressed by IFNγ-producing
CD8 + T cells, CD4 + T cells, as well as other cell types. _TIM3_ is a negative immunological checkpoint that is critical in the inhibition of the immune system induced by tumors. Another
research discovered that the transcription factor _TIM3_ was correlated with the immunosuppressive impact in HNSCC and that targeting _TIM3_ may boost anti-tumor immune reaction by
attenuating Tregs activity in HNSCC64. Tumor necrosis factor (_TNF_), consisting of _TNF-β_ and _TNF-α_, is mainly expressed on activated macrophages and lymphocytes. _TNF-α_ is also a
potent pro-inflammatory cytokine, which plays a critical role in the inflammatory response65. Because of its significant prognostic significance, _LAG3_ is often recognized as the most
important immunotherapy target inferior only to _PD-1_/_PDL-1_. _LAG3_ is not only expressed on naïve T cells, as is the case with _CTLA-4_ and _PD-1_, but also stimulated on CD8 + T and CD4
+ cells in response to antigen presentation. It cooperated with _PD-1_ to attenuate antitumor immunity and autoimmunity66. Lots of reports revealed the elevated expression of LAG3 and LAG3
+ cells infiltration in tumors is correlated with tumor progression, an unfavorable prognosis, as well as adverse clinical outcomes in a variety of human tumors, including colorectal
cancer67, renal cell carcinoma68 as well as HNSCC69. Additionally, the IPS analyses also illustrated that the low-risk score cohort exhibited a favorable clinical response to ICIs treatment,
which is logically consistent with the above ICGs analyses. Since TMB was demonstrated to be capable of predicting immune checkpoint blockade therapy efficacy and could be a useful
biomarker for the identification of patients that will benefit from immunotherapy across many cancer types70, we explored the topmost twenty commonly mutated genes in HNSCC and the
difference of mutation frequency in these genes. The results revealed _NOTCH1_ and _TP53_ mutations were highly frequent in the high-risk cohort while _CSMD3_, _PIK3CA_, _NSD1_, _USH2A_ had
obviously higher common mutations in the low-risk cohort. _CSMD3_ mutation and TME are rarely discussed by researchers, despite the fact that it is one of the most commonly mutated genes in
cancers. Several research reports have demonstrated that the _CSMD3_ mutation is correlated with favorable survival and one report introduced a case of high-grade serous ovarian cancer with
_CSMD3_ mutated exhibited a long-term response to anti-PD1 without prior chemotherapy71. _NSD1_ mutation is also suggested to be a favorable biomarker correlated with favorable survival in
HPV negative HNSCC, and its loss-of-function increases sensitivity to cisplatin72. Additionally, _PIK3CA_, _TP53_, and _NOTCH1_ mutation are well known associated with aberrant activation of
oncogenic pathways73. Overall, our analysis is consistent with the previous study, except that in our study, _PIK3CA_ exhibited highly frequent mutations in the low-risk cohort, on the
contrary, the underlying mechanism may need further exploration. Moreover, our result confirmed the prognosis predictive effect of the rrlncRNAs signature in high and low TMB cohorts.
CONCLUSION In conclusion, the ten-redox-related lncRNAs signature showed an excellent prognosis predictive value in HNSCC. Furthermore, the rrlncRNAs signature was associated with tumor
immune landscape in HNSCC. Moreover, this signature might indicate a response to chemotherapy and immunotherapy in HNSCC. DATA AVAILABILITY The datasets generated during the current study
are available in the public database for The Cancer Genome Atlas (TCGA) program, which is publicly available at https://portal.gdc.cancer.gov. Correspondence and requests for materials
should be addressed to M.T. REFERENCES * Johnson, D. E. _et al._ Head and neck squamous cell carcinoma. _Nat. Rev. Dis. Prim._ https://doi.org/10.1038/s41572-020-00224-3 (2020). Article
Google Scholar * Ferlay, J. _et al._ Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. _Int. J. Cancer_ 144, 1941–1953.
https://doi.org/10.1002/ijc.31937 (2019). Article CAS Google Scholar * Zhang, L. W. _et al._ Incidence and mortality trends in oral and oropharyngeal cancers in China, 2005–2013. _Cancer
Epidemiol._ 57, 120–126. https://doi.org/10.1016/j.canep.2018.10.014 (2018). Article Google Scholar * Blot, W. _et al._ Smoking and drinking in relation to oral and pharyngeal cancer.
_Can. Res._ 48, 3282–3287 (1988). CAS Google Scholar * Cramer, J. D., Burtness, B. & Ferris, R. L. Immunotherapy for head and neck cancer: Recent advances and future directions. _Oral
Oncol._ 99, 104460. https://doi.org/10.1016/j.oraloncology.2019.104460 (2019). Article CAS Google Scholar * Ferris, R. L. Immunology and immunotherapy of head and neck cancer. _J. Clin.
Oncol._ 33, 3293–3304. https://doi.org/10.1200/JCO.2015.61.1509 (2015). Article CAS Google Scholar * Zhang, Y. & Zhang, Z. The history and advances in cancer immunotherapy:
Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. _Cell Mol. Immunol._ 17, 807–821. https://doi.org/10.1038/s41423-020-0488-6 (2020).
Article CAS Google Scholar * Burtness, B. _et al._ Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the
head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. _Lancet_ 394, 1915–1928. https://doi.org/10.1016/S0140-6736(19)32591-7 (2019). Article CAS Google Scholar * Ferris,
R. L. _et al._ Nivolumab for recurrent squamous-cell carcinoma of the head and neck. _N. Engl. J. Med._ 375, 1856–1867. https://doi.org/10.1056/NEJMoa1602252 (2016). Article CAS Google
Scholar * Pulte, D. & Brenner, H. Changes in survival in head and neck cancers in the late 20th and early 21st century: A period analysis. _Oncologist_ 15, 994–1001.
https://doi.org/10.1634/theoncologist.2009-0289 (2010). Article Google Scholar * Kumpitsch, C., Moissl-Eichinger, C., Pock, J., Thurnher, D. & Wolf, A. Preliminary insights into the
impact of primary radiochemotherapy on the salivary microbiome in head and neck squamous cell carcinoma. _Sci. Rep._ 10, 16582. https://doi.org/10.1038/s41598-020-73515-0 (2020). Article
CAS Google Scholar * Swartz, J. _et al._ Poor prognosis in human papillomavirus-positive oropharyngeal squamous cell carcinomas that overexpress hypoxia inducible factor-1α. _Head Neck_
38, 1338–1346. https://doi.org/10.1002/hed.24445 (2016). Article Google Scholar * Florean, C., Song, S., Dicato, M. & Diederich, M. Redox biology of regulated cell death in cancer: A
focus on necroptosis and ferroptosis. _Free Radical Biol. Med._ 134, 177–189. https://doi.org/10.1016/j.freeradbiomed.2019.01.008 (2019). Article CAS Google Scholar * Helfinger, V. &
Schroder, K. Redox control in cancer development and progression. _Mol. Aspects Med._ 63, 88–98. https://doi.org/10.1016/j.mam.2018.02.003 (2018). Article CAS Google Scholar * Sharma, A.,
Rajappa, M., Saxena, A. & Sharma, M. Antioxidant status in advanced cervical cancer patients undergoing neoadjuvant chemoradiation. _Br. J. Biomed. Sci._ 64, 23–27.
https://doi.org/10.1080/09674845.2007.11732751 (2007). Article CAS Google Scholar * Sharma, A., Rajappa, M., Satyam, A. & Sharma, M. Oxidant/anti-oxidant dynamics in patients with
advanced cervical cancer: Correlation with treatment response. _Mol. Cell. Biochem._ 341, 65–72. https://doi.org/10.1007/s11010-010-0437-2 (2010). Article CAS Google Scholar * Gupta, A.
_et al._ Oxidative stress in non-small cell lung cancer patients after chemotherapy: Association with treatment response. _Respirology_ 15, 349–356.
https://doi.org/10.1111/j.1440-1843.2009.01703.x (2010). Article Google Scholar * Xiang, N., Zhao, R. & Zhong, W. Sodium selenite induces apoptosis by generation of superoxide via the
mitochondrial-dependent pathway in human prostate cancer cells. _Cancer Chemother. Pharmacol._ 63, 351–362. https://doi.org/10.1007/s00280-008-0745-3 (2009). Article CAS Google Scholar *
Mercer, T., Dinger, M. & Mattick, J. Long non-coding RNAs: Insights into functions. _Nat. Rev. Genet._ 10, 155–159. https://doi.org/10.1038/nrg2521 (2009). Article CAS Google Scholar
* Statello, L., Guo, C. J., Chen, L. L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. _Nat. Rev. Mol. Cell Biol._ 22, 96–118.
https://doi.org/10.1038/s41580-020-00315-9 (2021). Article CAS Google Scholar * Tang, Y., Li, C., Zhang, Y. J. & Wu, Z. H. Ferroptosis-related long non-coding RNA signature predicts
the prognosis of head and neck squamous cell carcinoma. _Int. J. Biol. Sci._ 17, 702–711. https://doi.org/10.7150/ijbs.55552 (2021). Article CAS Google Scholar * Li, H. _et al._
Establishment of a novel ferroptosis-related lncRNA pair prognostic model in colon adenocarcinoma. _Aging_ https://doi.org/10.18632/aging.203599 (2021). Article Google Scholar * Qi-Dong,
X. _et al._ Development and validation of a nine-redox-related long noncoding RNA signature in renal clear cell carcinoma. _Oxid. Med. Cell. Longev._ 2020, 6634247.
https://doi.org/10.1155/2020/6634247 (2020). Article CAS Google Scholar * Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. _Nucleic Acids Res._ 28, 27–30.
https://doi.org/10.1093/nar/28.1.27 (2000). Article CAS Google Scholar * Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis
of pathways and genomes. _Nucleic Acids Res._ https://doi.org/10.1093/nar/gkac963 (2022). Article Google Scholar * Yoshihara, K. _et al._ Inferring tumour purity and stromal and immune
cell admixture from expression data. _Nat. Commun._ 4, 2612. https://doi.org/10.1038/ncomms3612 (2013). Article ADS CAS Google Scholar * Danilova, L. _et al._ Programmed cell death
ligand-1 (PD-L1) and CD8 expression profiling identify an immunologic subtype of pancreatic ductal adenocarcinomas with favorable survival. _Cancer Immunol. Res._ 7, 886–895.
https://doi.org/10.1158/2326-6066.cir-18-0822 (2019). Article CAS Google Scholar * Charoentong, P. _et al._ Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships
and predictors of response to checkpoint blockade. _Cell Rep._ 18, 248–262. https://doi.org/10.1016/j.celrep.2016.12.019 (2017). Article CAS Google Scholar * Chalmers, Z. _et al._
Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. _Genome Med._ 9, 34. https://doi.org/10.1186/s13073-017-0424-2 (2017). Article CAS Google Scholar
* Marur, S. & Forastiere, A. A. Head and neck squamous cell carcinoma: Update on epidemiology, diagnosis, and treatment. _Mayo Clin. Proc._ 91, 386–396.
https://doi.org/10.1016/j.mayocp.2015.12.017 (2016). Article Google Scholar * Aggarwal, N. _et al._ Human papillomavirus infection in head and neck squamous cell carcinomas:
Transcriptional triggers and changed disease patterns. _Front. Cell. Infect. Microbiol._ 10, 537650. https://doi.org/10.3389/fcimb.2020.537650 (2020). Article CAS Google Scholar * Chen,
X. _et al._ Modulators of redox metabolism in head and neck cancer. _Antioxid. Redox Signal._ 29, 1660–1690. https://doi.org/10.1089/ars.2017.7423 (2018). Article CAS Google Scholar *
Huang, G. & Pan, S. T. ROS-Mediated therapeutic strategy in chemo-/radiotherapy of head and neck cancer. _Oxid. Med. Cell. Longev._ 2020, 5047987. https://doi.org/10.1155/2020/5047987
(2020). Article Google Scholar * Shen, Y., Peng, X. & Shen, C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. _Genomics_ 112, 2640–2646.
https://doi.org/10.1016/j.ygeno.2020.02.015 (2020). Article CAS Google Scholar * Zhang, G. _et al._ Identification and potential mechanisms of a 4-lncRNA signature that predicts
prognosis in patients with laryngeal cancer. _Hum. Genomics_ 13, 36. https://doi.org/10.1186/s40246-019-0230-6 (2019). Article CAS Google Scholar * Loewen, G., Jayawickramarajah, J.,
Zhuo, Y. & Shan, B. Functions of lncRNA HOTAIR in lung cancer. _J. Hematol. Oncol._ 7, 90. https://doi.org/10.1186/s13045-014-0090-4 (2014). Article CAS Google Scholar * Luo, R.,
Song, J., Zhang, W. & Ran, L. Identification of MFI2-AS1, a novel pivotal lncRNA for prognosis of stage III/IV colorectal cancer. _Dig. Dis. Sci._ 65, 3538–3550.
https://doi.org/10.1007/s10620-020-06064-1 (2020). Article CAS Google Scholar * Guglas, K. _et al._ lncRNA in HNSCC: Challenges and potential. _Contemp. Oncol._ 21, 259–266.
https://doi.org/10.5114/wo.2017.72382 (2017). Article CAS Google Scholar * Dong, X., Yang, Z., Yang, H., Li, D. & Qiu, X. Long non-coding RNA MIR4435-2HG promotes colorectal cancer
proliferation and metastasis through miR-206/YAP1 axis. _Front. Oncol._ 10, 160. https://doi.org/10.3389/fonc.2020.00160 (2020). Article Google Scholar * Gao, L. F. _et al._ Inhibition of
MIR4435-2HG on invasion, migration, and EMT of gastric carcinoma cells by mediating MiR-138-5p/Sox4 axis. _Front. Oncol._ 11, 661288. https://doi.org/10.3389/fonc.2021.661288 (2021). Article
Google Scholar * Wang, S., Chen, X. & Qiao, T. Long non-coding RNA MIR4435-2HG promotes the progression of head and neck squamous cell carcinoma by regulating the miR-383-5p/RBM3
axis. _Oncol. Rep._ https://doi.org/10.3892/or.2021.8050 (2021). Article Google Scholar * Huang, J. _et al._ HBx-related long non-coding RNA DBH-AS1 promotes cell proliferation and
survival by activating MAPK signaling in hepatocellular carcinoma. _Oncotarget_ 6, 33791–33804. https://doi.org/10.18632/oncotarget.5667 (2015). Article Google Scholar * Muri, J. &
Kopf, M. Redox regulation of immunometabolism. _Nat. Rev. Immunol._ 21, 363–381. https://doi.org/10.1038/s41577-020-00478-8 (2021). Article CAS Google Scholar * Manda, G. _et al._ The
redox biology network in cancer pathophysiology and therapeutics. _Redox Biol._ 5, 347–357. https://doi.org/10.1016/j.redox.2015.06.014 (2015). Article CAS Google Scholar * Roberts, D.
D., Kaur, S. & Isenberg, J. S. Regulation of cellular redox signaling by matricellular proteins in vascular biology, immunology, and cancer. _Antioxid. Redox Signal._ 27, 874–911.
https://doi.org/10.1089/ars.2017.7140 (2017). Article CAS Google Scholar * Vene, R. _et al._ Redox remodeling allows and controls B-cell activation and differentiation. _Antioxid. Redox
Signal._ 13, 1145–1155. https://doi.org/10.1089/ars.2009.3078 (2010). Article CAS Google Scholar * Li, X. _et al._ Reactive oxygen species reprogram macrophages to suppress antitumor
immune response through the exosomal miR-155-5p/PD-L1 pathway. _J. Exp. Clin. Cancer Res._ 41, 41. https://doi.org/10.1186/s13046-022-02244-1 (2022). Article CAS Google Scholar * Zhao,
J., Bao, W. & Cai, W. Immune Infiltration landscape in lung squamous cell carcinoma implications. _Biomed. Res. Int._ 2020, 5981870. https://doi.org/10.1155/2020/5981870 (2020). Article
CAS Google Scholar * Zuo, S., Wei, M., Wang, S., Dong, J. & Wei, J. Pan-cancer analysis of immune cell infiltration identifies a prognostic immune-cell characteristic score (ICCS) in
lung adenocarcinoma. _Front. Immunol._ 11, 1218. https://doi.org/10.3389/fimmu.2020.01218 (2020). Article CAS Google Scholar * Sui, S. _et al._ An immune cell infiltration-based immune
score model predicts prognosis and chemotherapy effects in breast cancer. _Theranostics_ 10, 11938–11949. https://doi.org/10.7150/thno.49451 (2020). Article CAS Google Scholar * Ye, L.
_et al._ Tumor-infiltrating immune cells act as a marker for prognosis in colorectal cancer. _Front. Immunol._ 10, 2368. https://doi.org/10.3389/fimmu.2019.02368 (2019). Article CAS Google
Scholar * Zhou, D., Wang, J., Wang, J. & Liu, X. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of HNSCC with or without HPV infection.
_Am. J. Trans. Res._ 13, 2163–2180 (2021). CAS Google Scholar * Wright, C. A., Kozik, P., Zacharias, M. & Springer, S. Tapasin and other chaperones: Models of the MHC class I loading
complex. _Biol. Chem._ 385, 763–778. https://doi.org/10.1515/BC.2004.100 (2004). Article CAS Google Scholar * Stern, L. J., Potolicchio, I. & Santambrogio, L. MHC class II compartment
subtypes: Structure and function. _Curr. Opin. Immunol._ 18, 64–69. https://doi.org/10.1016/j.coi.2005.11.005 (2006). Article CAS Google Scholar * Sabbatino, F. _et al._ Role of human
leukocyte antigen system as a predictive biomarker for checkpoint-based immunotherapy in cancer patients. _Int. J. Mol. Sci._ https://doi.org/10.3390/ijms21197295 (2020). Article Google
Scholar * Rodig, S. _et al._ MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. _Sci. Trans. Med._
https://doi.org/10.1126/scitranslmed.aar3342 (2018). Article Google Scholar * Liu, X., Cheng, W., Li, H. & Song, Y. Identification and validation of cuproptosis-related LncRNA
signatures as a novel prognostic model for head and neck squamous cell cancer. _Cancer Cell Int._ 22, 345. https://doi.org/10.1186/s12935-022-02762-0 (2022). Article CAS Google Scholar *
Xiang, J. _et al._ A hypoxia-related lncRNA model for prediction of head and neck squamous cell carcinoma prognosis. _Cancer Med._ https://doi.org/10.1002/cam4.5102 (2022). Article Google
Scholar * Zhang, W., Xin, J., Lai, J. & Zhang, W. LncRNA LINC00184 promotes docetaxel resistance and immune escape via miR-105-5p/PD-L1 axis in prostate cancer. _Immunobiology_ 227,
152163. https://doi.org/10.1016/j.imbio.2021.152163 (2022). Article CAS Google Scholar * Lei, Q., Wang, D., Sun, K., Wang, L. & Zhang, Y. Resistance mechanisms of anti-PD1/PDL1
therapy in solid tumors. _Front. Cell Dev. Biol._ 8, 672. https://doi.org/10.3389/fcell.2020.00672 (2020). Article ADS Google Scholar * Zhai, L. _et al._ IDO1 in cancer: A gemini of
immune checkpoints. _Cell. Mol. Immunol._ 15, 447–457. https://doi.org/10.1038/cmi.2017.143 (2018). Article CAS Google Scholar * Liu, Y. _et al._ Blockade of IDO-kynurenine-AhR metabolic
circuitry abrogates IFN-gamma-induced immunologic dormancy of tumor-repopulating cells. _Nat. Commun._ 8, 15207. https://doi.org/10.1038/ncomms15207 (2017). Article ADS Google Scholar *
Van Allen, E. M. _et al._ Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. _Science_ 350, 207–211. https://doi.org/10.1126/science.aad0095 (2015). Article ADS CAS
Google Scholar * Liu, J. F. _et al._ Blockade of TIM3 relieves immunosuppression through reducing regulatory T cells in head and neck cancer. _J. Exp. Clin. Cancer Res._ 37, 44.
https://doi.org/10.1186/s13046-018-0713-7 (2018). Article CAS Google Scholar * Horiuchi, T., Mitoma, H., Harashima, S., Tsukamoto, H. & Shimoda, T. Transmembrane TNF-alpha: structure,
function and interaction with anti-TNF agents. _Rheumatology_ 49, 1215–1228. https://doi.org/10.1093/rheumatology/keq031 (2010). Article CAS Google Scholar * Maruhashi, T., Sugiura, D.,
Okazaki, I. M. & Okazaki, T. LAG-3: From molecular functions to clinical applications. _J. Immunoth. Cancer_ https://doi.org/10.1136/jitc-2020-001014 (2020). Article Google Scholar *
Chen, J. & Chen, Z. The effect of immune microenvironment on the progression and prognosis of colorectal cancer. _Med. Oncol._ 31, 82. https://doi.org/10.1007/s12032-014-0082-9 (2014).
Article CAS Google Scholar * Giraldo, N. A. _et al._ Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer.
_Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res._ 21, 3031–3040. https://doi.org/10.1158/1078-0432.CCR-14-2926 (2015). Article CAS Google Scholar * Deng, W. _et al._ LAG-3 confers poor
prognosis and its blockade reshapes antitumor response in head and neck squamous cell carcinoma. _Oncoimmunology_ 5, e1239005. https://doi.org/10.1080/2162402x.2016.1239005 (2016). Article
Google Scholar * Chan, T. A. _et al._ Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. _Annal. Oncol. Off. J. Eur. Soc. Med. Oncol._ 30,
44–56. https://doi.org/10.1093/annonc/mdy495 (2019). Article CAS Google Scholar * Terzic, J. _et al._ Sustained response to pembrolizumab without prior chemotherapy in high-grade serous
ovarian carcinoma with CSMD3 mutation. _Gynecol. Oncol. Rep._ 33, 100600. https://doi.org/10.1016/j.gore.2020.100600 (2020). Article Google Scholar * Bui, N. _et al._ NSD1Disruption of in
head and neck cancer promotes favorable chemotherapeutic responses linked to hypomethylation. _Mol. Cancer Ther._ 17, 1585–1594. https://doi.org/10.1158/1535-7163.mct-17-0937 (2018). Article
CAS Google Scholar * Cancer Genome Atlas Network Genome sequencing centre. Comprehensive genomic characterization of head and neck squamous cell carcinomas. _Nature_ 517, 576–582.
https://doi.org/10.1038/nature14129 (2015). Article ADS CAS Google Scholar Download references ACKNOWLEDGEMENTS We express our gratitude to the TCGA program and to all of the R
programming package developers. FUNDING The Ningbo Technology Innovation 2025 Major Special Project (No. 2020Z097 and No. 2018B10015) supported this work. The Ningbo Public Welfare Science
and Technology fund (No. 2021S171). AUTHOR INFORMATION Author notes * These authors contributed equally: Kaitai Liu and Tianyi Huang. AUTHORS AND AFFILIATIONS * Department of Radiation
Oncology, The Lihuili Hospital, Ningbo Medical Center, Ningbo, Zhejiang, China Kaitai Liu, Tianyi Huang & Hui Zhang * Department of Otorhinolaryngology Head and Neck Surgery, The Lihuili
Hospital, Ningbo Medical Center, Ningbo, Zhejiang, China Hongxia Deng * Department of Otorhinolaryngology Head and Neck Surgery, Ningbo Women and Children’s Hospital, Ningbo, Zhejiang,
China Ming Tang Authors * Kaitai Liu View author publications You can also search for this author inPubMed Google Scholar * Tianyi Huang View author publications You can also search for this
author inPubMed Google Scholar * Hui Zhang View author publications You can also search for this author inPubMed Google Scholar * Hongxia Deng View author publications You can also search
for this author inPubMed Google Scholar * Ming Tang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.T. and K.T.L. collaborated on the
design of the study. K.T.L. and H.X.D. conducted data analyses from online databases. T.Y.H., H.Z. and H.X.D. were involved in the data analysis. H.Z. and K.T.L. was in charge of drafting
the manuscript. The manuscript was approved by all of the authors. CORRESPONDING AUTHOR Correspondence to Ming Tang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION 1. SUPPLEMENTARY INFORMATION 2. SUPPLEMENTARY INFORMATION 3. SUPPLEMENTARY INFORMATION 4. SUPPLEMENTARY INFORMATION 5. RIGHTS AND PERMISSIONS OPEN
ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format,
as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third
party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the
article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the
copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Liu, K., Huang, T., Zhang,
H. _et al._ Establishment and validation of a redox-related long non-coding RNAs prognostic signature in head and neck squamous cell carcinoma. _Sci Rep_ 12, 22040 (2022).
https://doi.org/10.1038/s41598-022-26490-7 Download citation * Received: 21 August 2022 * Accepted: 15 December 2022 * Published: 21 December 2022 * DOI:
https://doi.org/10.1038/s41598-022-26490-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Anti-vaxxers pose an immediate, serious threat to global public health. It's time to get tough. | thearticleThere is no more fertile breeding ground for conspiracy theories than social media. Perhaps the most dangerous of these ...
Our decisions and actions over what is now a very short period of time will determine how habitable our planet is in decades and generations to come —Due to Covid, this is my first time in the USA in three years, and so it is an absolute pleasure to be in Washington aga...
John swinney’s keynote address to conference #snp24 — scottish national partyCONFERENCE, IT IS AN IMMENSE PRIVILEGE FOR ME TO ADDRESS YOU TODAY – FOR THE FIRST TIME – AS SCOTLAND’S FIRST MINISTER....
Fbi files: how nazi spy ring was exposed by a double agent in ww2The FBI report details how “before America ever fired a shot” in World War 2, they uncovered a massive spy ring. The wor...
The page you were looking for doesn't exist.You may have mistyped the address or the page may have moved.By proceeding, you agree to our Terms & Conditions and our ...
Latests News
Establishment and validation of a redox-related long non-coding rnas prognostic signature in head and neck squamous cell carcinomaABSTRACT Reduction and oxidation (redox) reactions occur in living organisms as part of normal cellular metabolism. Here...
3 Surprising Things You Never Knew About the Post Office1:03 Videos de AARP 3 Surprising Things You Never Knew About the Post Office Facebook Twitter LinkedIn The U.S. Postal S...
Medical expense tax deduction: key characteristics by stateToday taxpayers with high out-of-pocket medical expenses may be able to deduct a portion of their costs from their taxab...
The page you were looking for doesn't exist.You may have mistyped the address or the page may have moved.By proceeding, you agree to our Terms & Conditions and our ...
Reuse DVSA learning materials - GOV.UKGuidance REUSE DVSA LEARNING MATERIALS How to get permission from the Driver and Vehicle Standards Agency (DVSA) to reus...