A descriptive analysis of antimicrobial resistance patterns of who priority pathogens isolated in children from a tertiary care hospital in india
A descriptive analysis of antimicrobial resistance patterns of who priority pathogens isolated in children from a tertiary care hospital in india"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The World Health Organization (WHO) has articulated a priority pathogens list (PPL) to provide strategic direction to research and develop new antimicrobials. Antimicrobial
resistance (AMR) patterns of WHO PPL in a tertiary health care facility in Southern India were explored to understand the local priority pathogens. Culture reports of laboratory specimens
collected between 1st January 2014 and 31st October 2019 from paediatric patients were extracted. The antimicrobial susceptibility patterns for selected antimicrobials on the WHO PPL were
analysed and reported. Of 12,256 culture specimens screened, 2335 (19%) showed culture positivity, of which 1556 (66.6%) were organisms from the WHO-PPL. _E. coli_ was the most common
organism isolated (37%), followed by _Staphylococcus aureus_ (16%). Total of 72% of _E. coli_ were extended-spectrum beta-lactamases (ESBL) producers, 55% of _Enterobacteriaceae_ were
resistant to 3rd generation cephalosporins due to ESBL, and 53% of _Staph. aureus_ were Methicillin-resistant. The analysis showed AMR trends and prevalence patterns in the study setting and
the WHO-PPL document are not fully comparable. This kind of local priority difference needs to be recognised in local policies and practices. SIMILAR CONTENT BEING VIEWED BY OTHERS
CEFTRIAXONE RESISTANCE AMONG PATIENTS AT GAMBY TEACHING GENERAL HOSPITAL Article Open access 14 July 2022 MAJOR BACTERIAL ISOLATE AND ANTIBIOTIC RESISTANCE FROM ROUTINE CLINICAL SAMPLES IN
SOUTHERN ETHIOPIA Article Open access 05 October 2021 BACTERIAL INFECTIONS EPIDEMIOLOGY AND FACTORS ASSOCIATED WITH MULTIDRUG RESISTANCE IN THE NORTHERN REGION OF GHANA Article Open access
21 December 2022 INTRODUCTION Antimicrobial resistance (AMR) has been recognised as a major threat to global health1. According to the World Health Organization (WHO), mutations in
microorganisms resulting in AMR, which consequently render medicines ineffective and infections persist in the body, increasing the risk of spread to others1. There are many reasons behind
the development of AMR, ranging from microbial causes to human aspects such as overuse and over-prescription of antimicrobials, agricultural and commercial application of antimicrobials in
the animal sector, and human behavioural factors2. Our ability to treat common pathogens becomes challenging because of AMR, resulting in increased duration of illness, costs, number of
complications, and deaths. By 2050, an estimated 10 million deaths are projected to occur due to AMR3, while another study projected AMR to cost the global economy US$100 trillion, in the
same period4. In 2015, the 68th World Health Assembly endorsed the Global Action Plan on AMR to tackle this global challenge5. This action plan has five strategic actions, focusing on (1)
improving awareness and understanding of AMR; (2) strengthening AMR surveillance; (3) reducing the incidence of infections; (4) optimizing antimicrobial use; and (5) developing the economic
case for AMR control. To support the Global Action Plan, WHO has developed a priority pathogens list (PPL), through a consultative process6. The prioritization process involved
multi-criteria decision analysis (MCDA) which used information from multiple sources, including disease mortality, transmissibility, treatability, health care burden, preventability in
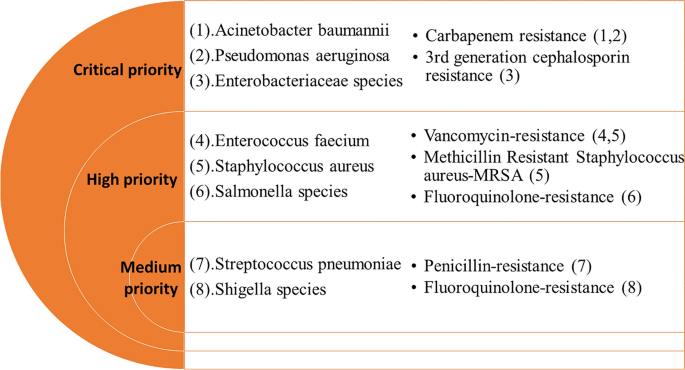
health care settings, and preventability in community settings, etc. Twelve families of drug-resistant bacteria, posing the greatest threat to human health, were categorized as critical,
high, and medium priority organisms, in terms of their resistance to selected antimicrobials (Fig. 1). Although this categorization was intended to prioritize and stimulate research and
develop new antimicrobials for specific drug resistance, it also makes a call for the prevention of infection and the rational use of antibiotics in both humans and animals6. Indian
population is known to be the highest consumer of antibiotics in the world7. The AMR situation in India has raised grave public health concerns8 and an action plan for its control is
considered crucial9,10. Given its importance for human health, the Government of India has developed a National Action Plan on Antimicrobial Resistance (NAP-AMR) 2017–202111. Strengthening
the knowledge and evidence base through surveillance of AMR is one of the five key strategies of this action plan. The Indian Council of Medical Research (ICMR) has established an
Antimicrobial Resistance Surveillance & Research Network (AMRSN) across selected hospitals in India, focusing on drug resistance among six pathogens12. However, not many hospitals
outside this network in India track AMR patterns among these pathogens. Generating AMR related evidence from a larger number of hospitals is critical for informed decision making on AMR
related policies and practices at local settings. This study explores the AMR susceptibility patterns for WHO priority pathogens identified in clinical isolates collected in the Paediatrics
Department of a tertiary care hospital in Southern India. The results of the culture tests are mainly availed for treatment purposes but are not systematically analysed on a routine basis.
The analysis of culture results could provide further evidence and guidance for the development of antimicrobial resistance control policy at the hospital and elsewhere. This analysis aims
to compare AMR patterns in WHO priority pathogens identified in a tertiary health care facility to understand the local priorities that can be applied to local policies and practices.
RESULTS A total of 12,256 culture specimens collected at paediatrics outpatient department and paediatrics inpatient wards were screened for bacteriological results, of which 2335 (19%)
showed culture positivity. Of these, 1556 were from the set of WHO PPL organisms. The largest number of bacterial isolation was seen in urine specimens (755/1556) followed by blood
(241/1556) (Table 1). _E. coli_ was the most common organism isolated (576), followed by _Staphylococcus aureus_ (252). Among the main WHO PPL organisms identified, 72% of _E. coli_ and 63%
of _Klebsiella_ spp. were resistant to 3rd generation cephalosporins due to extended-spectrum beta-lactamase (ESBL), and 53% of the _Staph. aureus_ were Methicillin-resistant (Table 2).
Overall, nearly half of _Enterobacteriaceae_ were resistant to carbapenem (46%) or 3rd generation cephalosporins due to ESBL (55%). The carbapenem resistance in _Pseudomonas aeruginosa_ was
found low (5%). Time trend analysis of selected WHO ‘Critical priority’ pathogens over the past 4 years showed a high proportion of resistance for carbapenem in _E coli_, _Klebsiella
pneumoniae,_ and _Enterobacter cloacae_ (Fig. 2). Similarly, _E. coli_ and _Klebsiella pneumoniae_ continued to show a high proportion of ESBL (Fig. 3). Among the WHO ‘High priority’
pathogens, _Staph. aureus_ continued to show a high proportion of methicillin-resistance (Fig. 4). Three of the WHO ‘High priority’ pathogens namely, _Helicobacter pylori_ (clarithromycin-
resistant), _Campylobacter_ spp. (fluoroquinolone-resistant), and _Neisseria gonorrhoeae_ (3rd generation cephalosporin-resistant and fluoroquinolone-resistant), were not detected in our
study specimens and antimicrobial sensitivity information was not available. Similarly, one of the WHO ‘Medium priority’ pathogen, _Haemophilus influenzae_ (ampicillin-resistant), was not
observed in this study, and antimicrobial sensitivity information was not available. DISCUSSION The 2017 guidance document of WHO indicated the highest carbapenem resistance worldwide in
_Acinetobacter baumannii_ (91%) and _Pseudomonas aeruginosa_ (82%), which is one of the reasons for classifying them as Critical Priority6 pathogens. The same study reported > 50%
carbapenem resistance in _Acinetobacter baumannii_, and 31% to 50% carbapenem resistance in _Pseudomonas aeruginosa_ in the Indian sub-continent in the general population. Early results from
the surveillance data from up to 22 ICMR-AMRSN sites in India showed around 80% carbapenem resistance in _Acinetobacter baumannii_ and around 30% in _Pseudomonas aeruginosa_12. However,
another study in children from Mumbai, India, identified only 15% of _Pseudomonas aeruginosa_ are resistant to carbapenem13 which is comparable to current study. The carbapenem-resistance in
_Acinetobacter baumannii_ and _Pseudomonas aeruginosa_ are 12% and 5% respectively in the current study in paediatric population. The WHO report6 identified high carbapenem resistance in
_E. coli_ (55%), _Klebsiella_ (70%), and _Enterobacter_ spp. (59%) in the general population, which is not far apart to the findings in this study in paediatric population. The ICMR-AMRSN
data also showed similar carbapenem resistance prevalence in _Klebsiella pneumoniae_ (40–50%) but a significantly lower level in _E. coli_ (15–25%) in the general population12. The ESBL
trends in _E. coli_ and _Klebsiella_ spp. (70–80%) as well as methicillin-resistance in _Staph. aureus_ (53%) were high in this study and comparable to the WHO report. However, due to the
low sample size of _Salmonella_ spp., _Shigella_ spp. and _Streptococcus pneumoniae_, it may not be appropriate to compare the results from this study to others. Several studies on AMR have
been published in India in recent years13,14,15. A retrospective 5 year follow-up study in a tertiary care hospital in North India showed increasing trends of AMR in urinary tract
infection-causing isolates14. Increasing trends of AMR was observed among gram negative isolates from samples collected across seven hospitals in India over 4 years, but the reported
carbapenem resistance prevalence in _Klebsiella_ spp. (39%) and _E. coli_ (12%) were lower than current study15. Among _Enterobacteriaceae_ isolated from a paediatric tertiary care hospital
in Mumbai, 24% were extended spectrum beta-lactamase (ESBL) producers and 27% were carbapenem-resistant isolates showing a lower resistance level than the current study13. As our study is
based on a retrospective dataset, it has several limitations. Although the health facility maintains a good quality of clinical and laboratory services along with proper documentation, one
cannot ensure that the quality checks in retrospective data are fully compatible with the highest quality standards of a well-conducted prospective study. Although the sample collection,
microbiological analysis, and report updates use standard procedures, it has likely been conducted by different people over the 5 year period, which may have had inter-personnel variations
on the quality of laboratory procedures. It is possible that at any given point of the study period new laboratory staff may have joined, may have taken time while undergoing training to
implement standardized procedures. Also, laboratory reports are manually entered into computerized system which is subject to human error and specific terms used during data entry are
subject to human variations. There were no standard inclusion criteria for sample collection as it was generally left to the discretion of the treating physician. In conclusion, among the
WHO PPL pathogens, _E. coli, Klebsiella species_, _Enterobacteriaceae, and Staph. aureus_ (methicillin-resistant) have high AMR in the study site. On the other hand, AMR patterns for
_Acinetobacter baumanni_, _Pseudomonas aeruginosa,_ and _Staph. aureus_ (vancomycin resistant) is lower than the WHO global estimates. These findings can guide local priorities, policy, and
practices. We recommend large health facilities to monitor and review emerging AMR patterns and trends periodically to prioritise, plan, and implement health facility level policies and
guidelines for the optimal use of antimicrobials. METHODS The study was conducted at the Yenepoya Medical College Hospital in Mangalore, South India. Typically, the Paediatric Department
collected around 2000 clinical specimens every year for culture tests from both outpatient and hospitalized cases. The sources and types of specimens collected for culture included blood,
urine, stool, pus, cerebrospinal fluid (CSF), sputum, and any other bodily fluids or other clinical specimens such as catheter, umbilical, and central line tips. As this is a retrospective
study, the sample selection for specimen collection was left to the discretion of the treating physician as sampling criteria was not predefined. The specimens were referred to the
laboratory in the Department of Microbiology for antimicrobial culture tests and antibiogram. The Kirby-Bauer disk diffusion method and/or by BD phoenix automated system were used for
performing antimicrobial susceptibility patterns and reported according to standard (Clinical Laboratory Standards Institute—CLSI) guidelines16,17,18,19,20. The confirmation of ESBL was done
as per the same CLSI guidelines. Once tests were performed, results were entered into the computer backbone system at the Department of Microbiology which was a specific database for the
tertiary hospital included in the study. The antibiogram reports generally covered the following antimicrobials: Carbapenem, Chloramphenicol, Cotrimoxazole, Nitrofurantoin, Piperacillin,
Piperacillin-Tazobactam, Tetracyclin, Tigecycline, Aztreonam, Amikacin, Gentamycin, Tobramycin, Ciprofloxacin, Levofloxacin, Norfloxacin, Cefoperazone, Ceftriaxone, Cefotaxime, Ceftazidime,
Cefepime, Cefazolin, Cefuroxime, Cefoxitin, Imipenem, Meropenem, Polymyxin B, and Colistin. The sample collection, microbiological analysis, and report entry on the computer were done on a
routine basis alongside the provision of healthcare services. These test results for antimicrobial susceptibility patterns were retrospectively accessed through the computer backbone system
during this study. Administrative permission to access laboratory culture records was obtained from Yenepoya Medical College. Retrospective culture reports between 1st January 2014 to 31st
October 2019 from various clinical specimens were extracted from the computer backbone system. The culture access numbers for all specimens with positive results were used to track the
antibiogram (i.e., which antibiotics were tested, and which were susceptible or resistant) results. The antibiogram for all culture isolates was extracted. Culture access numbers were also
used to track the culture source and the date of sample collection to the antibiograms. Culture reports of all paediatric cases were included in the study, irrespective of the location of
sample collection, namely: outpatients, inpatient wards, and neonatal and paediatric intensive care units (ICUs). The laboratory reports indicating contamination were excluded at data entry.
The reports containing duplicate or repeat samples, from the same source and subject, were also excluded so that the results for the same pathogens are not duplicated or repeated in the
analysis. Pathogens other than those in the WHO PPL were excluded from the analysis. The antimicrobial sensitivity tests other than the ones listed in WHO PPL were also excluded from the
analysis. Some of the WHO PPL pathogens were not included in this study as culture specimens were collected from children which do not generally include genitourinary swabs or gastric biopsy
specimens suitable for the culture of _Neisseria gonorrhoeae_ or _Helicobacter pylori_. Similarly, _Campylobacter_ spp. was also not identified in the specimens either because of low
incidence or because samples collected may not be best suited for its isolation. The data entry and analysis were performed on Microsoft Excel. The data from the computer backbone was
entered directly on a master Excel sheet followed by the removal of duplicates. De-identified data were organised by specimen types (such as blood, urine, etc.) in chronological order of
specimen collection date. A list of WHO PPL bacterial pathogens isolated were prepared by their species (such as _E coli_), specimen type, and resistance patterns. The list classified
pathogens into three main groups (Critical Priority, High Priority, and Medium Priority) for selected antimicrobials, based on WHO PPL (Fig. 1). The PPL defines the priority of pathogens
based on resistance to specific antimicrobials such as carbapenems, 3rd generation cephalosporins, vancomycin, methicillin, penicillins, or fluoroquinolones. A table presenting the number of
organisms isolated in the study site by specimen type was prepared. Another table was prepared to present selected AMR patterns in specific pathogens as defined by WHO PPL. Time trend
graphs were prepared for some key pathogens. ETHICAL ISSUES This study did not involve human subjects directly. An approval from the scientific and ethics committees of Yenepoya Medical
College (Name: _Yenepoya Ethics Committee-1_) was obtained for this study. The _Yenepoya Ethics Committee-1_ waived the need for participants to provide informed consent. To maintain
confidentiality, no identifiable information such as names, addresses, or phone numbers of subjects were collected. The data set, once finalised, was delinked from culture access numbers
before analysis, to retain confidentiality. ETHICS APPROVAL AND CONSENT TO PARTICIPATE The _Yenepoya Ethics Committee-1_ waived the need for participants to provide informed consent as
described under the manuscript. This study did not involve human subjects directly, no consent process was involved. All methods were carried out in accordance with relevant guidelines and
regulations. CONSENT FOR PUBLICATION All authors have consented for publication. DATA AVAILABILITY All the data included in the manuscript. REFERENCES * WHO. Antimicrobial resistance2018
October 13, 2019. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 13 Oct 2019. * Michael, C. A., Dominey-Howes, D. & Labbate, M. The antimicrobial
resistance crisis: Causes, consequences, and management. _Front. Public Health._ 2, 145 (2014). Article PubMed PubMed Central Google Scholar * Trotter, A. J., Aydin, A., Strinden, M. J.
& O’Grady, J. Recent and emerging technologies for the rapid diagnosis of infection and antimicrobial resistance. _Curr. Opin. Microbiol._ 51, 39–45 (2019). Article CAS PubMed Google
Scholar * O’Neill J. Review on Antimicrobial Resistance. Tackling drug Resistant Infections Globally: Final Report and Recommendations2016 October 13, 2019.
https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf. Accessed 13 Oct 2019. * WHO. Global action plan on antimicrobial resistance2015 October 13, 2019.
http://www.who.int/antimicrobial-resistance/global-action-plan/en/. Accessed 13 Oct 2019. * WHO. Prioritization of pathogens to guide discovery, research and development of new antibiotics
for drug resistant bacterial infections, including tuberculosis 2017 October 13, 2019. https://www.who.int/medicines/areas/rational_use/prioritization-of-pathogens/en/. Accessed 13 Oct 2019.
* Laxminarayan, R. & Chaudhury, R. R. Antibiotic resistance in india: Drivers and opportunities for action. _PLoS Med._ 13(3), e1001974 (2016). Article PubMed PubMed Central Google
Scholar * Ganguly N. Situation Analysis: Antibiotic Use and Resistance in India 2011 October 13, 2019. https://cddep.org/wp-content/uploads/2017/06/india-report-web_8.pdf. Accessed 13 Oct
2019. * Kumar, S. G. _et al._ Antimicrobial resistance in India: A review. _J. Nat. Sci. Biol. Med._ 4(2), 286–291 (2013). Article PubMed PubMed Central Google Scholar * Kakkar, M.,
Walia, K., Vong, S., Chatterjee, P. & Sharma, A. Antibiotic resistance and its containment in India. _BMJ_ 358, j2687 (2017). Article PubMed Google Scholar * National Action Plan on
Antimicrobial Resistance (NAP-AMR) 2017–2021. http://www.searo.who.int/india/topics/antimicrobial_resistance/nap_amr.pdf. Accessed 13 Oct 2019. * Walia, K. _et al._ Establishing
antimicrobial resistance surveillance & research network in india: Journey so far. _Indian J. Med. Res._ 149(2), 164–179 (2019). Article PubMed PubMed Central Google Scholar *
Thacker, N. _et al._ Epidemiology of blood stream infections in pediatric patients at a Tertiary Care Cancer Centre. _Indian J. Cancer._ 51(4), 438–441 (2014). Article CAS PubMed Google
Scholar * Patwardhan, V., Kumar, D., Goel, V. & Singh, S. Changing prevalence and antibiotic drug resistance pattern of pathogens seen in community-acquired pediatric urinary tract
infections at a tertiary care hospital of North India. _J. Lab. Physicians._ 9(4), 264–268 (2017). Article CAS PubMed PubMed Central Google Scholar * Veeraraghavan, B. _et al._
Antimicrobial susceptibility profiles of gram-negative bacteria causing infections collected across India during 2014–2016: Study for monitoring antimicrobial resistance trend report.
_Indian J. Med. Microbiol._ 36(1), 32–36 (2018). Article PubMed Google Scholar * CLSI-M100-24-2014. Performance Standards for Antimicrobial Susceptibility Testing (M100). 24th ed. The
Clinical Laboratory Standards Institute 2014. https://clsi.org/standards/products/microbiology/documents/m100/. Accessed 12 Dec 2020. * CLSI-M100-25-2015. Performance Standards for
Antimicrobial Susceptibility Testing (M100). 25th ed. The Clinical Laboratory Standards Institute 2015. https://clsi.org/standards/products/microbiology/documents/m100/. Accessed 12 Dec
2020. * CLSI-M100-26-2016. Performance Standards for Antimicrobial Susceptibility Testing (M100). 26th ed. The Clinical Laboratory Standards Institute 2016.
https://clsi.org/standards/products/microbiology/documents/m100/. Accessed 12 Dec 2020. * CLSI-M100-27-2017. Performance Standards for Antimicrobial Susceptibility Testing (M100). 27th ed.
The Clinical Laboratory Standards Institute 2017. https://clsi.org/standards/products/microbiology/documents/m100/. Accessed 12 Dec 2020. * CLSI-M100-28-2018. Performance Standards for
Antimicrobial Susceptibility Testing (M100). 28th ed. The Clinical Laboratory Standards Institute 2018. https://clsi.org/standards/products/microbiology/documents/. Accessed 9 Mar 2020.
Download references ACKNOWLEDGEMENTS We thank Mr. Satyajit Sarkar for editing and Ms. Athira Ramesh for research support. FUNDING This research was funded by the International Vaccine
Institute by the Swedish International Development Cooperation Agency [5410054] and the Bill & Melinda Gates Foundation. The International Vaccine Institute acknowledges its donors
including the Republic of Korea and the Republic of India. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Paediatrics, Yenepoya Medical College, Mangalore, India Vijayalaxmi V.
Mogasale & Prakash Saldanha * Department of Microbiology, Yenepoya Medical College, Mangalore, India Vidya Pai * Yenepoya Research Centre, Yenepoya (Deemed to be) University, Mangalore,
India P. D. Rekha * Policy and Economic Research Department, International Vaccine Institute, Seoul, South Korea Vittal Mogasale Authors * Vijayalaxmi V. Mogasale View author publications
You can also search for this author inPubMed Google Scholar * Prakash Saldanha View author publications You can also search for this author inPubMed Google Scholar * Vidya Pai View author
publications You can also search for this author inPubMed Google Scholar * P. D. Rekha View author publications You can also search for this author inPubMed Google Scholar * Vittal Mogasale
View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS V.V.M. served as Principal Investigator for this study and took overall responsibility for
protocol development, study design, ethics approval, data collection, analysis, and drafted the manuscript. V.M. conceptualized the study design, guided data extraction, analysis, and edited
the manuscript. P.S., V.S., and R.P.D. provided institutional support in data collection, expert advice, and contributed to the manuscript. All authors have approved the final version of
the manuscript. CORRESPONDING AUTHOR Correspondence to Vittal Mogasale. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION
PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS OPEN ACCESS This article
is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in
this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's
Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Mogasale, V.V., Saldanha, P., Pai, V. _et
al._ A descriptive analysis of antimicrobial resistance patterns of WHO priority pathogens isolated in children from a tertiary care hospital in India. _Sci Rep_ 11, 5116 (2021).
https://doi.org/10.1038/s41598-021-84293-8 Download citation * Received: 19 July 2020 * Accepted: 09 February 2021 * Published: 04 March 2021 * DOI:
https://doi.org/10.1038/s41598-021-84293-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Error 404Error 404 No encontramos la página que buscas....
Banks of another kind: these help dehradun manage its plastic waste efficientlyImagine 2600 trucks laden with plastic waste plying across cities, depositing them at waste processing centres to the ex...
3, 4, 5%? : what drop in french property prices is expected this year?A PRICE FALL IS ‘THE LAST AND ONLY LEVER’ TO RESTART THE DECLINING PROPERTY MARKET, SAY INDUSTRY EXPERTS Property prices...
9 questions to ask yourself before accepting a job offerMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...
M mot v iss facility services ltd: 2212308/2023M MOT V ISS FACILITY SERVICES LTD: 2212308/2023 Employment Tribunal decision. Read the full decision in M Mot v ISS Faci...
Latests News
A descriptive analysis of antimicrobial resistance patterns of who priority pathogens isolated in children from a tertiary care hospital in indiaABSTRACT The World Health Organization (WHO) has articulated a priority pathogens list (PPL) to provide strategic direct...
Banks hand out £2bn in bounce back loans in 24 hoursRishi Sunak revealed that the seven largest lenders – Barclays, Danske, HSBC, Lloyds, RBS, Santander and Virgin Money – ...
The page you were looking for doesn't exist.You may have mistyped the address or the page may have moved.By proceeding, you agree to our Terms & Conditions and our ...
Council leader on "bitter blow" which will lead to social care chargesCONTROVERSIAL MEANS-TESTED FEES FOR NON-RESIDENTIAL CARE IN EASTRENFREWSHIRE ARE ON THE HORIZON, WRITES LOCAL DEMOCRACY ...
Criticism of edweek's aft story (opinion)There’s been some interesting pushback against EdWeek’s story about the declining influence of the AFT -- not all of it ...