Chromatin integration labeling for mapping dna-binding proteins and modifications with low input
Chromatin integration labeling for mapping dna-binding proteins and modifications with low input"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
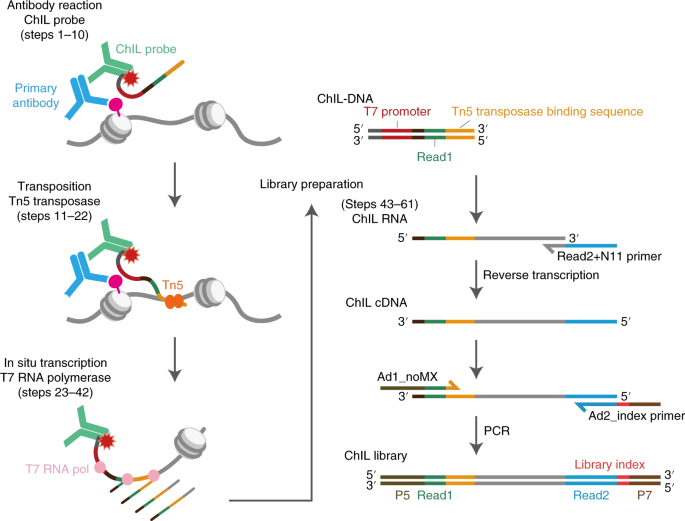
ABSTRACT Cell identity is determined by the selective activation or silencing of specific genes via transcription factor binding and epigenetic modifications on the genome. Chromatin
immunoprecipitation (ChIP) has been the standard technique for mapping the sites of transcription factor binding and histone modification. Recently, alternative methods to ChIP have been
developed for addressing the increasing demands for low-input epigenomic profiling. Chromatin integration labeling (ChIL) followed by sequencing (ChIL-seq) has been demonstrated to be
particularly useful for epigenomic profiling of low-input samples or even single cells because the technique amplifies the target genomic sequence before cell lysis. After labeling the
target protein or modification in situ with an oligonucleotide-conjugated antibody (ChIL probe), the nearby genome sequence is amplified by Tn5 transposase-mediated transposition followed by
T7 RNA polymerase-mediated transcription. ChIL-seq enables the detection of the antibody target localization under a fluorescence microscope and at the genomic level. Here we describe the
detailed protocol of ChIL-seq with assessment methods for the key steps, including ChIL probe reaction, transposition, in situ transcription and sequencing library preparation. The protocol
usually takes 3 d to prepare the sequencing library, including overnight incubations for the ChIL probe reaction and in situ transcription. The ChIL probe can be separately prepared and
stored for several months, and its preparation and evaluation protocols are also documented in detail. An optional analysis for multiple targets (multitarget ChIL-seq) is also described. We
anticipate that the protocol presented here will make the ChIL technique more widely accessible for analyzing precious samples and facilitate further applications. Access through your
institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio
journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per
year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated
during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MULTIPLEXED
CHROMATIN IMMUNOPRECIPITATION SEQUENCING FOR QUANTITATIVE STUDY OF HISTONE MODIFICATIONS AND CHROMATIN FACTORS Article 03 October 2024 SCNANOSEQ-CUT&TAG: A SINGLE-CELL LONG-READ
CUT&TAG SEQUENCING METHOD FOR EFFICIENT CHROMATIN MODIFICATION PROFILING WITHIN INDIVIDUAL CELLS Article 07 October 2024 EFFICIENT LOW-COST CHROMATIN PROFILING WITH CUT&TAG Article
10 September 2020 DATA AVAILABILITY ChIL-seq and mtChIL-seq data in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE140659. Source data are
provided with this paper. CODE AVAILABILITY All performance metrics of low-input epigenomic profiling methods in Table 2 are the re-evaluation of our original ChIL-seq data analysis (except
for the adapted information from previous reports, specified in the table legend). The main workhorse is the tableTPFP function in RScripts/myROC.R in
https://github.com/kazumits/DataAnalysisForChILT. The use case of calculating ROC at TSS is according to the example described at
https://github.com/kazumits/DataAnalysisForChILT/blob/master/ChILT_optimalResolution.md. The evaluation of genome-wide prediction performance (for example, precision, recall) against
gold-standard ChIP-seq peaks is shown at https://github.com/kazumits/DataAnalysisForChILT/blob/master/ChILT_sensitivity-specificity.md. Source data are provided with this paper. CHANGE
HISTORY * _ 29 NOVEMBER 2023 A Correction to this paper has been published: https://doi.org/10.1038/s41596-023-00940-6 _ REFERENCES * Bannister, A. J. & Kouzarides, T. Regulation of
chromatin by histone modifications. _Cell Res._ 21, 381–395 (2011). Article CAS PubMed PubMed Central Google Scholar * Kimura, H. Histone modifications for human epigenome analysis. _J.
Hum. Genet._ 58, 439–445 (2013). Article CAS PubMed Google Scholar * Harada, A. et al. A chromatin integration labelling method enables epigenomic profiling with lower input. _Nat. Cell
Biol._ 21, 287–296 (2019). Article CAS PubMed Google Scholar * Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. _Nat. Commun._
10, 1930 (2019). Article PubMed PubMed Central Google Scholar * Carter, B. et al. Mapping histone modifications in low cell number and single cells using antibody-guided chromatin
tagmentation (ACT-seq). _Nat. Commun._ 10, 3747 (2019). Article PubMed PubMed Central Google Scholar * Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. _Mol.
Cell_ 76, 206–216.e207 (2019). Article CAS PubMed Google Scholar * Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low
cell numbers. _Nat. Protoc._ 13, 1006–1019 (2018). Article CAS PubMed Google Scholar * Ku, W. L. et al. Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone
modification. _Nat. Methods_ 16, 323–325 (2019). Article CAS PubMed PubMed Central Google Scholar * Ai, S. et al. Profiling chromatin states using single-cell itChIP-seq. _Nat. Cell
Biol._ 21, 1164–1172 (2019). Article CAS PubMed Google Scholar * Zhu, B. et al. MOWChIP-seq for low-input and multiplexed profiling of genome-wide histone modifications. _Nat. Protoc._
14, 3366–3394 (2019). Article CAS PubMed PubMed Central Google Scholar * Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. _Science_ 345, 943–949
(2014). Article CAS PubMed PubMed Central Google Scholar * Dahl, J. A. et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. _Nature_ 537,
548–552 (2016). Article CAS PubMed PubMed Central Google Scholar * Brind’Amour, J. et al. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations.
_Nat. Commun._ 6, 6033 (2015). Article PubMed Google Scholar * Zhang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. _Nature_ 537,
553–557 (2016). Article CAS PubMed Google Scholar * van Galen, P. et al. A multiplexed system for quantitative comparisons of chromatin landscapes. _Mol. Cell_ 61, 170–180 (2016).
Article PubMed Google Scholar * Cao, Z., Chen, C., He, B., Tan, K. & Lu, C. A microfluidic device for epigenomic profiling using 100 cells. _Nat. Methods_ 12, 959–962 (2015). Article
CAS PubMed PubMed Central Google Scholar * Hainer, S. J., Boskovic, A., McCannell, K. N., Rando, O. J. & Fazzio, T. G. Profiling of pluripotency factors in single cells and early
embryos. _Cell_ 177, 1319–1329.e11 (2019). Article CAS PubMed PubMed Central Google Scholar * Rotem, A. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin
state. _Nat. Biotechnol._ 33, 1165–1172 (2015). Article CAS PubMed PubMed Central Google Scholar * Ma, S., Hsieh, Y. P., Ma, J. & Lu, C. Low-input and multiplexed microfluidic assay
reveals epigenomic variation across cerebellum and prefrontal cortex. _Sci. Adv._ 4, eaar8187 (2018). Article PubMed PubMed Central Google Scholar * Renaud, G., Stenzel, U., Maricic,
T., Wiebe, V. & Kelso, J. deML: robust demultiplexing of Illumina sequences using a likelihood-based approach. _Bioinformatics_ 31, 770–772 (2015). Article CAS PubMed Google Scholar
* Pombo, A. et al. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. _EMBO J._ 18, 2241–2253 (1999). Article CAS PubMed
PubMed Central Google Scholar * Kimura, H., Hayashi-Takanaka, Y., Goto, Y., Takizawa, N. & Nozaki, N. The organization of histone H3 modifications as revealed by a panel of specific
monoclonal antibodies. _Cell Struct. Funct._ 33, 61–73 (2008). Article CAS PubMed Google Scholar * Hayashi-Takanaka, Y. et al. Tracking epigenetic histone modifications in single cells
using Fab-based live endogenous modification labeling. _Nucleic Acids Res._ 39, 6475–6488 (2011). Article CAS PubMed PubMed Central Google Scholar * Chandra, T. et al. Independence of
repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. _Mol. Cell_ 47, 203–214 (2012). Article CAS PubMed PubMed Central Google Scholar *
Stasevich, T. J. et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. _Nature_ 516, 272–275 (2014). Article CAS PubMed Google Scholar *
Picelli, S. et al. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. _Genome Res._ 24, 2033–2040 (2014). Article CAS PubMed PubMed Central Google
Scholar * Sato, S. et al. Biochemical analysis of nucleosome targeting by Tn5 transposase. _Open Biol._ 9, 190116 (2019). Article CAS PubMed PubMed Central Google Scholar Download
references ACKNOWLEDGEMENTS We are grateful to the staff of Ohkawa Lab. The computation was carried out using the computer resource offered under the category of Intensively Promoted
Projects by Research Institute for Information Technology, Kyushu University. This work was in part supported by MEXT/JSPS KAKENHI (JP17K17719 to T.H.; JP18K19432, JP19H03211, JP19H05425 and
JP20H05368 to A.H.; JP18H04904, JP19H04970, JP19H03158 and JP20H05393 to K.M.; JP18H05534 to H.Ku.; JP18H04802, JP18H05527, JP19H05244, JP17H03608, JP20H00456, JP20H04846 and JP20K21398 to
Y.O.; and JP18H05527 and JP17H01417 to H.Ki), JST CREST (JPMJCR16G1 to Y.O., H.Ku. and H.Ki.), JST PRESTO JPMJPR19K7 to A.H, AMED JP20ek0109489h0001 to Y.O. and AMED BINDS (JP19am0101076 to
H.Ku. and JP19am0101105 to H.Ki.). AUTHOR INFORMATION Author notes * These authors contributed equally: Tetsuya Handa, Akihito Harada, Kazumitsu Maehara. AUTHORS AND AFFILIATIONS * Cell
Biology Center, Institute of Innovative Research, Tokyo Institute of Technology, 4259 Nagatsuta, Midori-ku, Yokohama, Japan Tetsuya Handa & Hiroshi Kimura * Division of Transcriptomics,
Medical Institute of Bioregulation, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka, Japan Akihito Harada, Kazumitsu Maehara & Yasuyuki Ohkawa * Institute for Quantitative
Biosciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo, Japan Shoko Sato & Hitoshi Kurumizaka * Graduate School of Bioscience and Biotechnology, Tokyo Institute of
Technology, 4259 Nagatsuta, Midori-ku, Yokohama, Japan Masaru Nakao, Naoki Goto & Hiroshi Kimura Authors * Tetsuya Handa View author publications You can also search for this author
inPubMed Google Scholar * Akihito Harada View author publications You can also search for this author inPubMed Google Scholar * Kazumitsu Maehara View author publications You can also search
for this author inPubMed Google Scholar * Shoko Sato View author publications You can also search for this author inPubMed Google Scholar * Masaru Nakao View author publications You can
also search for this author inPubMed Google Scholar * Naoki Goto View author publications You can also search for this author inPubMed Google Scholar * Hitoshi Kurumizaka View author
publications You can also search for this author inPubMed Google Scholar * Yasuyuki Ohkawa View author publications You can also search for this author inPubMed Google Scholar * Hiroshi
Kimura View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS T.H., A.H., Y.O. and H. Ki. designed the experiments. T.H. and A.H. performed the
ChIL experiments with deep-sequencing analyses. K.M. performed the bioinformatics and statistical analyses. S.S. and H.Ku. produced the in-house Tn5 transposase. M.N. and N.G. contributed to
the assay for in situ transcription. T.H., A.H., K.M., Y.O. and H.Ki. wrote the manuscript. All authors read and approved the final manuscript. CORRESPONDING AUTHORS Correspondence to
Yasuyuki Ohkawa or Hiroshi Kimura. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests except T.H., A.H., H. Ku., Y.O. and H.Ki, who are involved in
a pending patent related to ChIL. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Protocols_ thanks Chang Lu, Sinem K. Saka and the other, anonymous, reviewer(s) for their
contribution to the peer review of this work. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RELATED
LINKS KEY REFERENCE USING THIS PROTOCOL: Harada, A. et al. _Nat. Cell Biol_. 21, 287−296 (2019): https://doi.org/10.1038/s41556-018-0248-3 SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION
Supplementary Tables 1–5 and Figs. 1–9. REPORTING SUMMARY SOURCE DATA SOURCE DATA FIG. 2 Unprocessed gels. SOURCE DATA FIG. 3 Statistical source data. SOURCE DATA FIG. 4 Statistical source
data. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other
rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Handa, T., Harada, A., Maehara, K. _et al._ Chromatin integration labeling for mapping DNA-binding proteins and modifications with low input.
_Nat Protoc_ 15, 3334–3360 (2020). https://doi.org/10.1038/s41596-020-0375-8 Download citation * Received: 24 November 2019 * Accepted: 29 May 2020 * Published: 17 August 2020 * Issue Date:
October 2020 * DOI: https://doi.org/10.1038/s41596-020-0375-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Effect of laser fluence on the optoelectronic properties of nanostructured gan/porous silicon prepared by pulsed laser depositionABSTRACT In this study, the fabrication of nanostructured GaN/porous Si by pulsed laser deposition (PLD) was demonstrate...
Let's focus on gaps in opportunity, not achievement (opinion)Research tells us again and again that when students do not achieve, their underachievement is a function of the opportu...
How to stop snoring: doctor identifies easy way to improve sleepSNORING: DOCTOR EXPLAINS HOW TO SLEEP BETTER AT NIGHT Snoring is a very common issue that is caused when your tongue, mo...
President roosevelt paid a high price for america's global superiority | thearticlePresident Roosevelt is the father of American global superiority for three reasons. First, he gave the world freedom of ...
How to provide care for patients in eating disorder recoveryAccess through your institution Buy or subscribe Whilst in recovery for an eating disorder, I constantly struggled with ...
Latests News
Chromatin integration labeling for mapping dna-binding proteins and modifications with low inputABSTRACT Cell identity is determined by the selective activation or silencing of specific genes via transcription factor...
Mr s kelly v the zoltar group ltd and others: 2400708/2020MR S KELLY V THE ZOLTAR GROUP LTD AND OTHERS: 2400708/2020 Employment Tribunal decision. Read the full decision in Mr S ...
Effect of laser fluence on the optoelectronic properties of nanostructured gan/porous silicon prepared by pulsed laser depositionABSTRACT In this study, the fabrication of nanostructured GaN/porous Si by pulsed laser deposition (PLD) was demonstrate...
Hasan ali profile | hasan ali cricket career | cricket statsCNN name, logo and all associated elements ® and © 2024 Cable News Network LP, LLLP. A Time Warner Company. All rights r...
[withdrawn] construction and use of hydrogen-powered vehicles* Department for Transport Guidance CONSTRUCTION AND USE OF HYDROGEN-POWERED VEHICLES Published 1 April 2009 THIS GUIDAN...