Pi3kγ in b cells promotes antibody responses and generation of antibody-secreting cells
Pi3kγ in b cells promotes antibody responses and generation of antibody-secreting cells"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The differentiation of naive and memory B cells into antibody-secreting cells (ASCs) is a key feature of adaptive immunity. The requirement for phosphoinositide 3-kinase-delta
(PI3Kδ) to support B cell biology has been investigated intensively; however, specific functions of the related phosphoinositide 3-kinase-gamma (PI3Kγ) complex in B lineage cells have not.
In the present study, we report that PI3Kγ promotes robust antibody responses induced by T cell-dependent antigens. The inborn error of immunity caused by human deficiency in PI3Kγ results
in broad humoral defects, prompting our investigation of roles for this kinase in antibody responses. Using mouse immunization models, we found that PI3Kγ functions cell intrinsically within
activated B cells in a kinase activity-dependent manner to transduce signals required for the transcriptional program supporting differentiation of ASCs. Furthermore, ASC fate choice
coincides with upregulation of _PIK3CG_ expression and is impaired in the context of PI3Kγ disruption in naive B cells on in vitro CD40-/cytokine-driven activation, in memory B cells on
toll-like receptor activation, or in human tonsillar organoids. Taken together, our study uncovers a fundamental role for PI3Kγ in supporting humoral immunity by integrating signals
instructing commitment to the ASC fate. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this
journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS RIF1 INTEGRATES DNA REPAIR AND TRANSCRIPTIONAL REQUIREMENTS DURING THE ESTABLISHMENT OF HUMORAL IMMUNE RESPONSES Article Open access 17 January
2025 CONTROL OF GERMINAL CENTER B CELL SURVIVAL AND IGE PRODUCTION BY AN ADAPTOR MOLECULE CONTAINING PH AND SH2 DOMAINS, APS/SH2B2 Article Open access 01 August 2024 A P38Α-BLIMP1
SIGNALLING PATHWAY IS ESSENTIAL FOR PLASMA CELL DIFFERENTIATION Article Open access 28 November 2022 DATA AVAILABILITY Bulk RNA-seq and scRNA-seq and BCR-seq data are available via the Gene
Expression Omnibus (GEO) and were analyzed using mm10 reference genome. The bulk and scRNA-seq data used in the present study are available through the GEO database under accession nos.
GSE269303 and GSE269300, respectively. Source data are provided with this paper. CODE AVAILABILITY Customized scripts in R v.4.3.1 for scBCR analysis were deposited under
bitbucket.org/kleinstein/projects (Lanahan2023 folder). REFERENCES * Nutt, S. L., Hodgkin, P. D., Tarlinton, D. M. & Corcoran, L. M. The generation of antibody-secreting plasma cells.
_Nat. Rev. Immunol._ 15, 160–171 (2015). CAS PubMed Google Scholar * Brandtzaeg, P. et al. Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial
lymphocytes. _Gastroenterology_ 97, 1562–1584 (1989). CAS PubMed Google Scholar * van der Heijden, P. J., Stok, W. & Bianchi, A. T. Contribution of immunoglobulin-secreting cells in
the murine small intestine to the total ‘background’ immunoglobulin production. _Immunology_ 62, 551–555 (1987). PubMed PubMed Central Google Scholar * Gaudette, B. T. et al. Resting
innate-like B cells leverage sustained Notch2/mTORC1 signaling to achieve rapid and mitosis-independent plasma cell differentiation. _J. Clin. Invest._ 131_,_ e151975 (2021). *
Skrzypczynska, K. M., Zhu, J. W. & Weiss, A. Positive regulation of Lyn kinase by CD148 is required for B cell receptor signaling in B1 but not B2 B cells. _Immunity_ 45, 1232–1244
(2016). * Gaudette, B. T., Jones, D. D., Bortnick, A., Argon, Y. & Allman, D. mTORC1 coordinates an immediate unfolded protein response-related transcriptome in activated B cells
preceding antibody secretion. _Nat. Commun._ 11, 723 (2020). CAS PubMed PubMed Central Google Scholar * Tumang, J. R., Frances, R., Yeo, S. G. & Rothstein, T. L. Spontaneously
Ig-secreting B-1 cells violate the accepted paradigm for expression of differentiation-associated transcription factors. _J. Immunol._ 174, 3173–3177 (2005). CAS PubMed Google Scholar *
Martin, F., Oliver, A. M. & Kearney, J. F. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. _Immunity_ 14, 617–629 (2001).
CAS PubMed Google Scholar * Klein, U. et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. _Nat. Immunol._ 7, 773–782 (2006). CAS PubMed
Google Scholar * Shaffer, A. L. et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. _Immunity_ 17, 51–62 (2002). CAS
PubMed Google Scholar * Carotta, S. et al. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. _J. Exp. Med._ 211, 2169–2181 (2014). PubMed PubMed
Central Google Scholar * Scharer, C. D. et al. Antibody-secreting cell destiny emerges during the initial stages of B-cell activation. _Nat. Commun._ 11, 3989 (2020). CAS PubMed PubMed
Central Google Scholar * Jones, D. D. et al. mTOR has distinct functions in generating versus sustaining humoral immunity. _J. Clin. Invest._ 126, 4250–4261 (2016). PubMed PubMed Central
Google Scholar * Luo, W. et al. IL-21R signal reprogramming cooperates with CD40 and BCR signals to select and differentiate germinal center B cells. _Sci. Immunol._ 8, eadd1823 (2023).
CAS PubMed PubMed Central Google Scholar * Luo, W., Weisel, F. & Shlomchik, M. J. B cell receptor and CD40 signaling are rewired for synergistic induction of the c-Myc transcription
factor in germinal center B cells. _Immunity_ 48, 313–326.e315 (2018). CAS PubMed PubMed Central Google Scholar * Clayton, E. et al. A crucial role for the p110delta subunit of
phosphatidylinositol 3-kinase in B cell development and activation. _J. Exp. Med._ 196, 753–763 (2002). CAS PubMed PubMed Central Google Scholar * Conley, M. E. et al. Agammaglobulinemia
and absent B lineage cells in a patient lacking the p85alpha subunit of PI3K. _J. Exp. Med._ 209, 463–470 (2012). CAS PubMed PubMed Central Google Scholar * Coulter, T. I. et al.
Clinical spectrum and features of activated phosphoinositide 3-kinase delta syndrome: A large patient cohort study. _J. Allergy Clin. Immunol._ 139, 597–606.e594 (2017). CAS PubMed PubMed
Central Google Scholar * Bader, A. G., Kang, S., Zhao, L. & Vogt, P. K. Oncogenic PI3K deregulates transcription and translation. _Nat. Rev. Cancer_ 5, 921–929 (2005). CAS PubMed
Google Scholar * Vanhaesebroeck, B., Stephens, L. & Hawkins, P. PI3K signalling: the path to discovery and understanding. _Nat. Rev. Mol. Cell Biol._ 13, 195–203 (2012). CAS PubMed
Google Scholar * Burke, J. E. & Williams, R. L. Synergy in activating class I PI3Ks. _Trends Biochem. Sci._ 40, 88–100 (2015). CAS PubMed Google Scholar * Rommel, C., Camps, M. &
Ji, H. PI3Kδ and PI3Kγ: partners in crime in inflammation in rheumatoid arthritis and beyond? _Nat. Rev. Immunol._ 7, 191–201 (2007). CAS PubMed Google Scholar * Hirsch, E. et al.
Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. _Science_ 287, 1049–1053 (2000). CAS PubMed Google Scholar * Sasaki, T. et al. Function of PI3Kgamma in
thymocyte development, T cell activation, and neutrophil migration. _Science_ 287, 1040–1046 (2000). * Beer-Hammer, S. et al. The catalytic PI3K isoforms p110gamma and p110delta contribute
to B cell development and maintenance, transformation, and proliferation. _J. Leukoc. Biol._ 87, 1083–1095 (2010). CAS PubMed Google Scholar * Reif, K. et al. Cutting edge: differential
roles for phosphoinositide 3-kinases, p110gamma and p110delta, in lymphocyte chemotaxis and homing. _J. Immunol._ 173, 2236–2240 (2004). * Takeda, A. J. et al. Human PI3Kγ deficiency and its
microbiota-dependent mouse model reveal immunodeficiency and tissue immunopathology. _Nat. Commun._ 10, 4364 (2019). PubMed PubMed Central Google Scholar * Suire, S. et al. p84, a new
Gbetagamma-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110gamma. _Curr. Biol._ 15, 566–570 (2005). CAS PubMed Google Scholar * Stephens, L. R. et al. The G beta
gamma sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. _Cell_ 89, 105–114 (1997). CAS PubMed Google Scholar * Allen, D., Simon, T., Sablitzky, F., Rajewsky, K.
& Cumano, A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response. _EMBO J._ 7, 1995–2001 (1988). CAS PubMed PubMed Central Google Scholar *
Elsner, R. A. & Shlomchik, M. J. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. _Immunity_ 53, 1136–1150 (2020). CAS PubMed PubMed
Central Google Scholar * Casola, S. et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. _Proc. Natl Acad. Sci.
USA_ 103, 7396–7401 (2006). CAS PubMed PubMed Central Google Scholar * Breasson, L. et al. PI3Kγ activity in leukocytes promotes adipose tissue inflammation and early-onset insulin
resistance during obesity. _Sci. Signal_. 10, eaaf2969 (2017). * Nojima, T. et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. _Nat.
Commun._ 2, 465 (2011). PubMed Google Scholar * Monaco, G. et al. RNA-seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. _Cell Rep._ 26,
1627–1640 e1627 (2019). CAS PubMed PubMed Central Google Scholar * Lanahan, S. M., Wymann, M. P. & Lucas, C. L. The role of PI3Kγ in the immune system: new insights and translational
implications. _Nat. Rev. Immunol._ 22, 687–700 (2022). CAS PubMed PubMed Central Google Scholar * Heng, T. S., Painter, M. W. & Immunological Genome Project The Immunological Genome
Project: networks of gene expression in immune cells. _Nat. Immunol._ 9, 1091–1094 (2008). CAS PubMed Google Scholar * Pinto, D. et al. A functional BCR in human IgA and IgM plasma
cells. _Blood_ 121, 4110–4114 (2013). CAS PubMed Google Scholar * Wagar, L. E. et al. Modeling human adaptive immune responses with tonsil organoids. _Nat. Med._ 27, 125–135 (2021). CAS
PubMed PubMed Central Google Scholar * Le Coz, C. et al. Human T follicular helper clones seed the germinal center-resident regulatory pool. _Sci. Immunol._ 8, eade8162 (2023). PubMed
PubMed Central Google Scholar * Bekeredjian-Ding, I. & Jego, G. Toll-like receptors—sentries in the B-cell response. _Immunology_ 128, 311–323 (2009). CAS PubMed PubMed Central
Google Scholar * Gong, G. Q. et al. A small-molecule PI3Kα activator for cardioprotection and neuroregeneration. _Nature_ 618, 159–168 (2023). CAS PubMed PubMed Central Google Scholar *
Donahue, A. C. & Fruman, D. A. Distinct signaling mechanisms activate the target of rapamycin in response to different B-cell stimuli. _Eur. J. Immunol._ 37, 2923–2936 (2007). CAS
PubMed Google Scholar * Thian, M. et al. Germline biallelic PIK3CG mutations in a multifaceted immunodeficiency with immune dysregulation. _Haematologica_ 105, e448–e492 (2020). PubMed
Central Google Scholar * Camps, M. et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. _Nat. Med._ 11, 936–943 (2005). CAS
PubMed Google Scholar * Barber, D. F. et al. PI3Kgamma inhibition blocks glomerulonephritis and extends lifespan in a mouse model of systemic lupus. _Nat. Med._ 11, 933–935 (2005). CAS
PubMed Google Scholar * Del Prete, A. et al. Defective dendritic cell migration and activation of adaptive immunity in PI3Kγ-deficient mice. _EMBO J._ 23, 3505–3515 (2004). PubMed PubMed
Central Google Scholar * Nobs, S. P. et al. PI3Kγ is critical for dendritic cell-mediated CD8+ T cell priming and viral clearance during influenza virus infection. _PLoS Pathog._ 12,
e1005508 (2016). PubMed PubMed Central Google Scholar * Laidlaw, B. J. et al. The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. _J. Exp.
Med._ 214, 639–649 (2017). CAS PubMed PubMed Central Google Scholar * Rossbacher, J. & Shlomchik, M. J. The B cell receptor itself can activate complement to provide the complement
receptor 1/2 ligand required to enhance B cell immune responses in vivo. _J. Exp. Med._ 198, 591–602 (2003). CAS PubMed PubMed Central Google Scholar * Luo, W. et al. SREBP signaling is
essential for effective B cell responses. _Nat. Immunol._ 24, 337–348 (2023). CAS PubMed Google Scholar * Study to Evaluate the Efficacy/Safety of IPI-549 in Combination With Nivolumab in
Patients With Advanced Urothelial Carcinoma (MARIO-275)_. NIH_ https://clinicaltrials.gov/ct2/show/NCT03980041 (2019). * Evaluation of IPI-549 Combined With Front-line Treatments in Pts.
With Triple-Negative Breast Cancer or Renal Cell Carcinoma (MARIO-3)_. NIH_ https://clinicaltrials.gov/ct2/show/NCT03961698 (2019). * Evans, C. A. et al. Discovery of a selective
phosphoinositide-3-kinase (PI3K)-γ inhibitor (IPI-549) as an immuno-oncology clinical candidate. _ACS Med. Chem. Lett._ 7, 862–867 (2016). CAS PubMed PubMed Central Google Scholar *
Haniuda, K. & Kitamura, D. Induced germinal center B cell culture system. _Bio Protoc._ 9, e3163 (2019). PubMed PubMed Central Google Scholar * Pahl, M. C. et al. Implicating effector
genes at COVID-19 GWAS loci using promoter-focused Capture-C in disease-relevant immune cell types. _Genome Biol._ 23, 125 (2022). CAS PubMed PubMed Central Google Scholar * Ramaswamy,
A. et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. _Immunity_ 54, 1083–1095.e1087
(2021). CAS PubMed PubMed Central Google Scholar * Barmada, A. et al. Cytokinopathy with aberrant cytotoxic lymphocytes and profibrotic myeloid response in SARS-CoV-2 mRNA
vaccine-associated myocarditis. _Sci. Immunol._ 8, eadh3455 (2023). CAS PubMed PubMed Central Google Scholar * Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep
generative modeling for single-cell transcriptomics. _Nat. Methods_ 15, 1053–1058 (2018). CAS PubMed PubMed Central Google Scholar * Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY:
large-scale single-cell gene expression data analysis. _Genome Biol._ 19, 15 (2018). PubMed PubMed Central Google Scholar * Ye, J., Ma, N., Madden, T. L. & Ostell, J. M. IgBLAST: an
immunoglobulin variable domain sequence analysis tool. _Nucleic Acids Res._ 41, W34–40 (2013). PubMed PubMed Central Google Scholar * Lefranc, M. P. et al. IMGT, the international
ImMunoGeneTics information system. _Nucleic Acids Res._ 37, D1006–1012 (2009). CAS PubMed Google Scholar * Alsoussi, W. B. et al. A potently neutralizing antibody protects mice against
SARS-CoV-2 infection. _J. Immunol._ 205, 915–922 (2020). CAS PubMed Google Scholar * Gupta, N. T. et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire
sequencing data. _Bioinformatics_ 31, 3356–3358 (2015). CAS PubMed PubMed Central Google Scholar * Nouri, N. & Kleinstein, S. H. A spectral clustering-based method for identifying
clones from high-throughput B cell repertoire sequencing data. _Bioinformatics_ 34, i341–i349 (2018). CAS PubMed PubMed Central Google Scholar * Stern, J. N. et al. B cells populating
the multiple sclerosis brain mature in the draining cervical lymph nodes. _Sci. Transl. Med._ 6, 248ra107 (2014). PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS
We thank the patients and their families for participation in research and all clinical care staff for their contributions. We thank L. Wirth for technical assistance, J.-M. Carpier for
critical feedback and training, E. Fagerberg and J. Attanasio for contributions to RNA-seq experimental design and A. Rice for his contributions. We acknowledge D. Kitamura, Tokyo University
of Science, Organization for Research Advancement, Research Institute for Biomedical Sciences for the 40LB feeder cell line used in Extended Data Fig. 3. C.L.L. received funding for this
work from the Colton Center for Autoimmunity at Yale, NIH/NIAID (grant no. R21AI144315) and Yale University. M.P.W. was funded by the Swiss National Science Foundation (SNF; grant no.
310030_189065). N.R. is funded by the NIH, NIAID (grant no. AI146026 to N.R.), the Chan Zuckerberg Initiative Pediatric Human Cell Atlas and the Jeffrey Modell Foundation. BioRender.com was
used for schematics in Extended Data Fig. 2h and Extended Data Fig. 6. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Immunobiology, Yale University School of Medicine, New
Haven, CT, USA Stephen M. Lanahan, Lucas Yang, Kate M. Jones, Zhihong Qi, Anjali Ramaswamy, Anis Barmada, Dinesh Babu Uthaya Kumar, Lan Xu, Peiying Shan, Steven H. Kleinstein & Carrie L.
Lucas * Division of Immunology and Allergy, Children’s Hospital of Philadelphia, Philadelphia, PA, USA Emylette Cruz Cabrera & Neil Romberg * Immunology Graduate Group, Perelman School
of Medicine, Philadelphia, PA, USA Lauren Y. Cominsky * Department of Pathology, Yale University School of Medicine, New Haven, CT, USA Gisela Gabernet & Steven H. Kleinstein *
Department of Biomedicine, University of Basel, Basel, Switzerland Matthias P. Wymann * Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT, USA Steven H.
Kleinstein * Primary Immunodeficiency Clinic, Laboratory of Clinical Immunology and Microbiology, NIAID, NIH, Bethesda, MD, USA V. Koneti Rao * Division of Allergy and Immunology, Department
of Pediatrics, Nationwide Children’s Hospital, Columbus, OH, USA Peter Mustillo * Department of Pediatrics, Perelman School of Medicine, Philadelphia, PA, USA Neil Romberg * Department of
Pathology and Laboratory Medicine, Nationwide Children’s Hospital, Columbus, OH, USA Roshini S. Abraham Authors * Stephen M. Lanahan View author publications You can also search for this
author inPubMed Google Scholar * Lucas Yang View author publications You can also search for this author inPubMed Google Scholar * Kate M. Jones View author publications You can also search
for this author inPubMed Google Scholar * Zhihong Qi View author publications You can also search for this author inPubMed Google Scholar * Emylette Cruz Cabrera View author publications You
can also search for this author inPubMed Google Scholar * Lauren Y. Cominsky View author publications You can also search for this author inPubMed Google Scholar * Anjali Ramaswamy View
author publications You can also search for this author inPubMed Google Scholar * Anis Barmada View author publications You can also search for this author inPubMed Google Scholar * Gisela
Gabernet View author publications You can also search for this author inPubMed Google Scholar * Dinesh Babu Uthaya Kumar View author publications You can also search for this author inPubMed
Google Scholar * Lan Xu View author publications You can also search for this author inPubMed Google Scholar * Peiying Shan View author publications You can also search for this author
inPubMed Google Scholar * Matthias P. Wymann View author publications You can also search for this author inPubMed Google Scholar * Steven H. Kleinstein View author publications You can also
search for this author inPubMed Google Scholar * V. Koneti Rao View author publications You can also search for this author inPubMed Google Scholar * Peter Mustillo View author publications
You can also search for this author inPubMed Google Scholar * Neil Romberg View author publications You can also search for this author inPubMed Google Scholar * Roshini S. Abraham View
author publications You can also search for this author inPubMed Google Scholar * Carrie L. Lucas View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS S.M.L. was involved in idea generation, performed experiments, analyzed data and wrote the manuscript. L.Y. and K.M.J. performed experiments, analyzed data and wrote methods
for their work. Z.Q., D.B.U.K., L.X. and P.S. conducted experiments and analyses. E.C.C., L.Y.C. and N.R. conducted and analyzed the tonsillar organoid experiments. A.R., A.B., G.G. and
S.H.K. performed analysis of single-cell transcriptome and BCR-seq. K.R., P.M. and R.A. provided patient care and analyses relating to patient A.1’s samples. M.P.W. provided KO and floxed
mice and input. C.L.L. supervised overall research and data analysis, was involved in idea generation and wrote and edited the manuscript. All authors discussed and reviewed the manuscript.
CORRESPONDING AUTHOR Correspondence to Carrie L. Lucas. ETHICS DECLARATIONS COMPETING INTERESTS C.L.L. reports an advisory/consulting role for Pharming Healthcare Inc. and unrelated funding
support from Ono Pharma. S.H.K. receives consulting fees from Peraton. R.S.A. discloses support from ClinGen, _Journal of Immunology_ and Clinical Laboratory Standards Institute. The
remaining authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Immunology_ thanks Sidonia Fagarasan and the other, anonymous, reviewer(s) for their
contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the
editorial team. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA
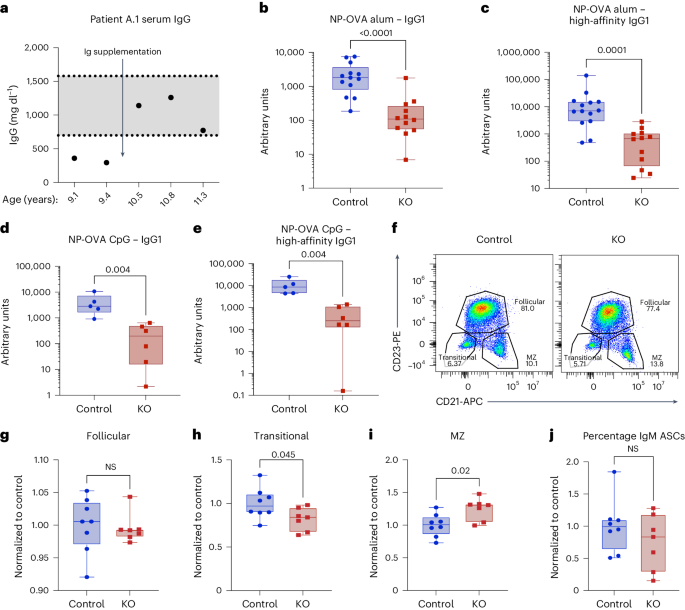
EXTENDED DATA FIG. 1 PI3KΓ IS REQUIRED FOR T-DEPENDENT ANTIBODY RESPONSES. (A-B) Serum IgM (A) and IgA (B) levels in Patient A.1. (C-E) Mice were immunized (as in Fig. 1b, c) and
antigen-specific IgM (day 5) (C), primary (non-boosted) IgG1 (day 12) (D), or IgG2c (E) was measured via high conjugation ratio NP-BSA ELISA. (F) Mice were immunized (as in Fig. 1d, e) and
IgG2b was measured via NP-BSA ELISA. Data are from 2-3 independent experiments and are presented as box-and-whisker plots showing median, min, and max. Data points represent different
biological samples. Statistical analysis was performed using non-parametric two-tailed unpaired T-tests. (c) n = 12 for control, n = 12 for KO. (d) n = 15 for control, n = 14 for KO. (e) n =
12 for control, n = 11 for KO. (f) n = 5 for control, n = 6 for KO. Source data EXTENDED DATA FIG. 2 PI3KΓ IS REQUIRED FOR GC B CELLS TO ADOPT ASC GENE SIGNATURES. (A) Stacked bar plots
showing proportions of cell subsets within Naïve or GC B cells used for CSR/SHM analysis. (B) Percentage of high-affinity W38L mutations from the total number of sequences with IGHV1-72 gene
and lambda light chains. Statistical analysis was Wilcoxon Rank Sum (C-D) Quantification of marginal zone B cells (C) and follicular B cells (D) from the spleens of indicated mice assessed
by flow cytometry. Data are representative of 2 independent experiments (n = 5 for each condition). (E) Volcano plot showing gene expression from RNA sequencing in sorted naïve B cells (IgD+
B220+) from WT versus PI3Kγ KO mice 28 days after SRBC i.p. immunization. Data are from one experiment. (F) Volcano plot of differentially expressed genes from bulk RNA sequencing of sorted
GC B cells at day 28 post immunization, as described for Fig. 2e. (G) Expression of ‘Activated B cell’ UPR program genes in sorted splenic GC B cells from _Pik3cg_ heterozygous or KO mice
28 days after SRBC immunization. Transcriptional program is defined by Gaudette, et. al 2020. Data are from one experiment (n = 3 for het, n = 4 for KO). (H) Diagram of germinal center B
cells that are beginning to adopt expression of antibody secreting cell surface markers (CD138). Diagram was created with BioRender.com. (C-D) Each dot represents different biological
samples and data are presented as box-and-whisker plots showing median, min, and max. Statistical analysis was performed using non-parametric two-tailed unpaired T-tests. Source data
EXTENDED DATA FIG. 3 B CELLS LACKING PI3KΓ ACTIVATE NORMALLY AND ARE CAPABLE OF FORMING GERMINAL CENTERS AND NORMAL IGA RESPONSES. (A) Littermate control or conditional _Pik3cg_ KO mice were
immunized and boosted with NP-OVA precipitated in alum. Antigen-specific IgG2c (day 28) was measured via NP-BSA ELISA, and arbitrary units were defined using pooled immunized sera as a
standard. (B) Immunofluorescence imaging of spleens from indicated mice 28 days after SRBC immunization. Data are representative of two independent repeats (n = 6 for each genotype). (C-H)
Murine naïve B cells were stimulated using TI stimulation with TLR agonists (CpG or LPS) or TD stimulation with anti-CD40, anti-IgM F(ab’)2 and assessed for proliferation using cell trace
violet (gating on live cells after 4 days) and activation by staining for CD69 or CD86 (gating on live cells after 2 days). Data (n = 3 for each condition) are representative of two
independent experiments. (I-N) Quantification of B cell subsets from Peyer’s patches of mice assessed by flow cytometry. Data are representative of three independent experiments (n = 13 for
control and 14 for KO). (O) Quantification of fecal IgA in control versus B cell-specific PI3Kγ KO mice (n = 6 per group). (P) Quantification of surface IgG-expressing memory and GC B cells,
normalized to control animals, in control versus B cell-specific PI3Kγ KO mice (n = 3 for control and 5 for KO). (Q-R) _In-vitro_ generation of GC B cells using 40LB feeder cells, as in
Nojima et al. 2011. Data are from two independent experiments (n = 5 for each condition). Data are presented as a bar graph (median ± SD) or box-and-whisker plots showing median, min, and
max. Each dot represents different biological samples and statistical analysis was performed using non-parametric two-tailed unpaired T-tests, except for panels q-r which uses non-parametric
paired T-test. Source data EXTENDED DATA FIG. 4 FUNCTIONAL PI3KΓ IS REQUIRED FOR OPTIMAL ASC DIFFERENTIATION _IN VITRO_ AND _IN VIVO_ DESPITE BEING DISPENSABLE FOR INNATE-LIKE B CELL IGM
RESPONSES. (A) Expression of PI3K genes in human/mouse immune cells derived from Immgen3. (B) Expression of _Pik3cg_ in mouse B cells comparing splenic follicular and germinal center B cells
to innate-like marginal zone and B1a B cells. (C-D) Mice were immunized i.p. with TI antigens and serum was taken on day 5 to measure antigen-specific IgM via ELISA. (E-F) Antigen-specific
ASCs were measured from splenocytes via ELISPOT in WT mice treated as described for Fig. 4e, f. (G-H) Assessment of _in-vitro_ differentiation of naïve mouse B cells into ASCs (anti-CD40,
IL-4, IL-5 gating on CD138+ after 4 days) in utilizing B cells from Blimp1-YFP mice treated _in vitro_ with DMSO vehicle or the PI3Kγ inhibitor IPI-549. Data are from 2-4 independent
experiments. Data presented as box-and-whisker plots show median, min, and max. (c) n = 11 for controls, n = 12 for KOs. (d) n = 12 for both groups. (e) n = 15 for both groups. (f) n = 13
for both. (g-h) n = 6 for each group. Each dot represents different biological samples and statistical analysis was performed using non-parametric two-tailed unpaired T-tests, with the
exception of panel H which used non-parametric paired T-test. Source data EXTENDED DATA FIG. 5 PI3KΓ IS REQUIRED FOR ROBUST HUMAN ASC DIFFERENTIATION. (A-H) Cells from human tonsillar
organoid model were assessed via flow cytometry (n = 5). Graphs depict percentage of each subset or percentage of live cells in each subset. (I) Assessment of _in-vitro_ differentiation of
primary healthy human peripheral blood pan-B cells into IRF4+ cells with 100 nM dose of PI3Kγi IPI-549 versus DMSO (shown as a proportion) after 5 days in the presence of DMSO control or
PI3Kα activator (UCL-TRO-1938). (J-K) Overnight IPI-549 treatment of isolated primary human CD138+ ASCs from peripheral blood. ELISPOTs and quantification are from 2 independent experiments
(n = 6). Each dot represents different biological samples and data are presented as mean ± SD. Statistical analysis was performed using non-parametric two-tailed unpaired T-tests and paired
T-test for (i). Source data EXTENDED DATA FIG. 6 MODEL SUMMARIZING THE FUNCTION OF PI3KΓ IN ANTIBODY RESPONSES. B cell-intrinsic PI3Kγ supports the transition to the ASC transcriptional
program and ASC differentiation in activated B cells. Diagram was created with BioRender.com. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Patient A.1 antibody titers elicited by
indicated vaccines. REPORTING SUMMARY SOURCE DATA SOURCE DATA FIG. 1 Statistical source data. SOURCE DATA FIG. 2 Statistical source data. SOURCE DATA FIG. 3 Statistical source data. SOURCE
DATA FIG. 4 Statistical source data. SOURCE DATA FIG. 5 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 1 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 2 Statistical source
data. SOURCE DATA EXTENDED DATA FIG. 3 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 4 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 5 Statistical source data. RIGHTS
AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other
rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lanahan, S.M., Yang, L., Jones, K.M. _et al._ PI3Kγ in B cells promotes antibody responses and generation of antibody-secreting cells. _Nat
Immunol_ 25, 1422–1431 (2024). https://doi.org/10.1038/s41590-024-01890-1 Download citation * Received: 06 July 2023 * Accepted: 07 June 2024 * Published: 03 July 2024 * Issue Date: August
2024 * DOI: https://doi.org/10.1038/s41590-024-01890-1 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Soccer-bullet point preview of premier league fixtures, round 32Following are match-by-match facts and statistics ahead of round 32 of the Premier League fixtures on April 8-10 (1400 G...
France: stop your regular taxe d’habitation paymentSome 80% of main home households will have zero to pay on this year’s taxe d’habitation bill, which will be sent out in ...
Analysts reduce their inflation forecast—and growthPrivate analysts consulted by Mexico’s central bank have lowered their forecasts for both economic growth and inflation ...
Something went wrong, sorry. :(The Paris mairie is cracking down on ‘voyeurs’ and similar sexual assaults in its public swimming pools, after many wome...
allopen3 | designboom.com* imprint * privacy policy * terms of use * cookies * copyright info * contribute * about us * contact us * newsletter *...
Latests News
Pi3kγ in b cells promotes antibody responses and generation of antibody-secreting cellsABSTRACT The differentiation of naive and memory B cells into antibody-secreting cells (ASCs) is a key feature of adapti...
About the 2019 AARP Community ChallengeFacebook Twitter The application period for the 2019 AARP Community Challenge — a "quick-action" grant program to improv...
Hardline Hindus force interfaith Indian TV ad off airVideo IconVIDEOHardline Hindus force interfaith Indian TV ad off air...
Celtics, kristaps porzingis reportedly finalizing a two-year extensionCeltics THE DEAL WILL REPORTEDLY BE APPROXIMATELY WORTH $60 MILLION OVER THE TWO SEASONS. A little more than a week afte...
Indian leaders to share needs and concerns at white house tribal conferenceThis Thursday, President Obama will meet with Native American leaders from across the U.S. for the so-called White House...