Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis
Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
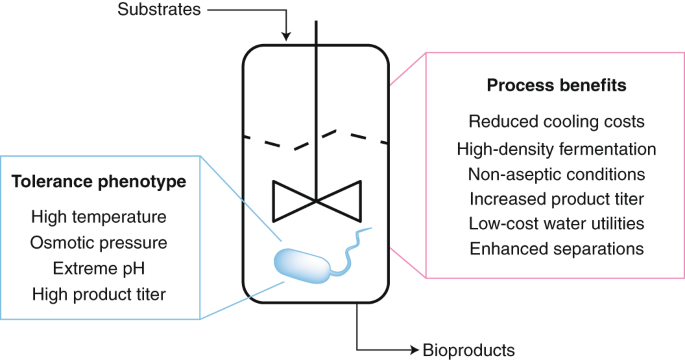
ABSTRACT Microbial chemical production is a rapidly growing industry, with much of the growth fueled by advances in synthetic biology. New approaches have enabled rapid strain engineering
for the production of various compounds; however, translation to industry is often problematic because native phenotypes of model hosts prevent the design of new low-cost bioprocesses. Here,
we argue for a new approach that leverages the native stress-tolerant phenotypes of non-conventional microbes that directly address design challenges from the outset. Growth at high
temperature, high salt and solvent concentrations, and low pH can enable cost savings by reducing the energy required for product separation, bioreactor cooling, and maintaining sterile
conditions. These phenotypes have the added benefit of allowing for the use of low-cost sugar and water resources. Non-conventional hosts are needed because these phenotypes are polygenic
and thus far have proven difficult to recapitulate in the common hosts _Escherichia coli_ and _Saccharomyces cerevisiae_. Access through your institution Buy or subscribe This is a preview
of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value
online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more
Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS:
* Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SCREENING NON-CONVENTIONAL YEASTS FOR ACID TOLERANCE AND
ENGINEERING _PICHIA OCCIDENTALIS_ FOR PRODUCTION OF MUCONIC ACID Article Open access 31 August 2023 PHYSIOLOGICAL LIMITATIONS AND OPPORTUNITIES IN MICROBIAL METABOLIC ENGINEERING Article 02
August 2021 GENOME-SCALE TARGET IDENTIFICATION IN _ESCHERICHIA COLI_ FOR HIGH-TITER PRODUCTION OF FREE FATTY ACIDS Article Open access 17 August 2021 REFERENCES * Carlson, R. Estimating the
biotech sector’s contribution to the US economy. _Nat. Biotechnol._ 34, 247–255 (2016). THIS PERSPECTIVE ARTICLE PROVIDES A DETAILED ANALYSIS OF THE US INDUSTRIAL BIOTECHNOLOGY SECTOR.
Article CAS PubMed Google Scholar * Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. _Science_ 337, 816–821 (2012). Article CAS PubMed
PubMed Central Google Scholar * Caruthers, M. H. A brief review of DNA and RNA chemical synthesis. _Biochem. Soc. Trans._ 39, 575–580 (2011). Article CAS PubMed Google Scholar * Chao,
R., Mishra, S., Si, T. & Zhao, H. Engineering biological systems using automated biofoundries. _Metab. Eng._ 42, 98–108 (2017). Article CAS PubMed PubMed Central Google Scholar *
Hong, K. K. & Nielsen, J. Metabolic engineering of _Saccharomyces cerevisiae_: a key cell factory platform for future biorefineries. _Cell. Mol. Life Sci._ 69, 2671–2690 (2012). Article
CAS PubMed Google Scholar * Pontrelli, S. et al. _Escherichia coli_ as a host for metabolic engineering. _Metab. Eng._ 50, 16–46 (2018). Article CAS PubMed Google Scholar *
Shapouri, H. & Gallagher, P. USDA’s 2002 Ethanol Cost-of-Production Survey. _Agricultural Economics Report Number 841_ (US Department of Agriculture, Office of Energy Policy and New
Uses, 2005). * Blanch, H.W. & Clark, D.S. _Biochemical Engineering_, xii (M. Dekker, 1996). * Lam, F. H., Ghaderi, A., Fink, G. R. & Stephanopoulos, G. Biofuels. Engineering alcohol
tolerance in yeast. _Science_ 346, 71–75 (2014). Article CAS PubMed PubMed Central Google Scholar * Rich, J. O., Leathers, T. D., Bischoff, K. M., Anderson, A. M. & Nunnally, M. S.
Biofilm formation and ethanol inhibition by bacterial contaminants of biofuel fermentation. _Bioresour. Technol._ 196, 347–354 (2015). Article CAS PubMed Google Scholar * Löbs, A. K.,
Engel, R., Schwartz, C., Flores, A. & Wheeldon, I. CRISPR–Cas9-enabled genetic disruptions for understanding ethanol and ethyl acetate biosynthesis in _Kluyveromyces marxianus_.
_Biotechnol. Biofuels_ 10, 164 (2017). Article PubMed PubMed Central CAS Google Scholar * Shui, W. et al. Understanding the mechanism of thermotolerance distinct from feat shock
response through proteomic analysis of industrial strains of _Saccharomyces cerevisiae_. _Mol. Cell. Proteom._ 14, 1885–1897 (2015). Article CAS Google Scholar * Richter, K., Haslbeck, M.
& Buchner, J. The heat shock response: life on the verge of death. _Mol. Cell_ 40, 253–266 (2010). Article CAS PubMed Google Scholar * Phipps, B. M. et al. Structure of a molecular
chaperone from a thermophilic Archaebacterium. _Nature_ 361, 475–477 (1993). Article CAS Google Scholar * Parsell, D. A. & Lindquist, S. The function of heat-shock proteins in stress
tolerance: degradation and reactivation of damaged proteins. _Annu. Rev. Genet._ 27, 437–496 (1993). Article CAS PubMed Google Scholar * Strassburg, K., Walther, D., Takahashi, H.,
Kanaya, S. & Kopka, J. Dynamic transcriptional and metabolic responses in yeast adapting to temperature stress. _OMICS_ 14, 249–259 (2010). Article CAS PubMed PubMed Central Google
Scholar * Balchin, D., Hayer-Hartl, M. & Hartl, F. U. In vivo aspects of protein folding and quality control. _Science_ 353, aac4354 (2016). Article PubMed CAS Google Scholar *
Verghese, J., Abrams, J., Wang, Y. & Morano, K. A. Biology of the heat shock response and protein chaperones: budding yeast (_Saccharomyces cerevisiae_) as a model system. _Microbiol.
Mol. Biol. Rev._ 76, 115–158 (2012). Article CAS PubMed PubMed Central Google Scholar * Gasch, A. P. et al. Genomic expression programs in the response of yeast cells to environmental
changes. _Mol. Biol. Cell_ 11, 4241–4257 (2000). Article CAS PubMed PubMed Central Google Scholar * Rodrigues, J. L. & Rodrigues, L. R. Potential applications of the _Escherichia
coli_ heat shock response in synthetic biology. _Trends Biotechnol._ 36, 186–198 (2018). Article CAS PubMed Google Scholar * Lewin, A., Wentzel, A. & Valla, S. Metagenomics of
microbial life in extreme temperature environments. _Curr. Opin. Biotechnol._ 24, 516–525 (2013). Article CAS PubMed Google Scholar * Jacquemet, A., Barbeau, J., Lemiègre, L. &
Benvegnu, T. Archaeal tetraether bipolar lipids: structures, functions and applications. _Biochimie_ 91, 711–717 (2009). Article CAS PubMed Google Scholar * Li, P. et al. The
transcription factors Hsf1 and Msn2 of thermotolerant _Kluyveromyces marxianus_ promote cell growth and ethanol fermentation of _Saccharomyces cerevisiae_ at high temperatures. _Biotechnol.
Biofuels_ 10, 289 (2017). Article PubMed PubMed Central CAS Google Scholar * Seong, Y. J. et al. Physiological and metabolomic analysis of _Issatchenkia orientalis_ MTY1 with multiple
tolerance for cellulosic bioethanol production. _Biotechnol. J._ 12, 1700110 (2017). Article CAS Google Scholar * Blaby, I. K. et al. Experimental evolution of a facultative thermophile
from a mesophilic ancestor. _Appl. Environ. Microbiol._ 78, 144–155 (2012). Article CAS PubMed PubMed Central Google Scholar * Rudolph, B., Gebendorfer, K. M., Buchner, J. & Winter,
J. Evolution of _Escherichia coli_ for growth at high temperatures. _J. Biol. Chem._ 285, 19029–19034 (2010). Article CAS PubMed PubMed Central Google Scholar * Caspeta, L. et al.
Biofuels. Altered sterol composition renders yeast thermotolerant. _Science_ 346, 75–78 (2014). Article CAS PubMed Google Scholar * Shi, D. J., Wang, C. L. & Wang, K. M. Genome
shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of _Saccharomyces cerevisiae_. _J. Ind. Microbiol. Biotechnol._ 36, 139–147 (2009). Article CAS PubMed
Google Scholar * Bhandiwad, A. et al. Metabolic engineering of _Thermoanaerobacterium saccharolyticum_ for n-butanol production. _Metab. Eng._ 21, 17–25 (2014). Article CAS PubMed Google
Scholar * Lin, P. P. et al. Consolidated bioprocessing of cellulose to isobutanol using _Clostridium thermocellum_. _Metab. Eng._ 31, 44–52 (2015). Article CAS PubMed Google Scholar *
Cripps, R. E. et al. Metabolic engineering of _Geobacillus thermoglucosidasius_ for high yield ethanol production. _Metab. Eng._ 11, 398–408 (2009). Article CAS PubMed Google Scholar *
Yue, H. T. et al. A seawater-based open and continuous process for polyhydroxyalkanoates production by recombinant _Halomonas campaniensis_ LS21 grown in mixed substrates. _Biotechnol.
Biofuels_ 7, 108 (2014). THIS PAPER SHOWS THE POWER OF STRESS-TOLERANT MICROBES. ENGINEERED HALOPHILIC _H. CAMPANIENSIS_ WAS USED IN NON-ASEPTIC BIOREACTORS TO PRODUCE HIGH TITERS OF
POLYHYDROXYBUTYRATE FROM MIXED SUBSTRATE FEEDSTOCKS. Article CAS Google Scholar * Voronovsky, A. Y., Rohulya, O. V., Abbas, C. A. & Sibirny, A. A. Development of strains of the
thermotolerant yeast _Hansenula polymorpha_ capable of alcoholic fermentation of starch and xylan. _Metab. Eng._ 11, 234–242 (2009). Article CAS PubMed Google Scholar * Löbs, A. K., Lin,
J. L., Cook, M. & Wheeldon, I. High throughput, colorimetric screening of microbial ester biosynthesis reveals high ethyl acetate production from Kluyveromyces marxianus on C5, C6, and
C12 carbon sources. _Biotechnol. J._ 11, 1274–1281 (2016). Article PubMed CAS Google Scholar * Xiao, H., Shao, Z., Jiang, Y., Dole, S. & Zhao, H. Exploiting _Issatchenkia orientalis_
SD108 for succinic acid production. _Microb. Cell Fact._ 13, 121 (2014). Article PubMed PubMed Central CAS Google Scholar * Löbs, A. K., Schwartz, C., Thorwall, S. & Wheeldon, I.
Highly multiplexed CRISPRi repression of respiratory functions enhances mitochondrial localized ethyl acetate biosynthesis in _Kluyveromyces marxianus_. _ACS Synth. Biol._ 7, 2647–2655
(2018). THIS PAPER DEMONSTRATES THE POWER OF CRISPR TECHNOLOGIES FOR RAPIDLY ENGINEERING NON-CONVENTIONAL HOSTS. HERE, MULTIPLEXED CRISPR INTERFERENCE (CRISPRI) WAS USED TO SCREEN VARIOUS
TRANSCRIPTIONAL PROGRAMS TO OPTIMIZE A NATIVE BIOSYNTHETIC PATHWAY IN THE THERMOTOLERANT YEAST _K. MARXIANUS_. Article PubMed CAS Google Scholar * Abdel-Banat, B. M., Hoshida, H., Ano,
A., Nonklang, S. & Akada, R. High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic
yeast? _Appl. Microbiol. Biotechnol._ 85, 861–867 (2010). Article CAS PubMed Google Scholar * Cernak, P. et al. Engineering _Kluyveromyces marxianus_ as a robust synthetic biology
platform host. _MBio_ 9, e01410–e01418 (2018). Article CAS PubMed PubMed Central Google Scholar * Cheon, Y. et al. A biosynthetic pathway for hexanoic acid production in _Kluyveromyces
marxianus_. _J. Biotechnol._ 182-183, 30–36 (2014). Article CAS PubMed Google Scholar * Dmytruk, K., Kurylenko, O., Ruchala, J., Ishchuk, O. & Sibirny, A. in _Yeast Diversity in
Human Welfare_ (eds Satyanarayana, T. & Kunze, G.) 257–282 (Springer, 2017). * Lenton, S., Walsh, D. L., Rhys, N. H., Soper, A. K. & Dougan, L. Structural evidence for
solvent-stabilisation by aspartic acid as a mechanism for halophilic protein stability in high salt concentrations. _Phys. Chem. Chem. Phys._ 18, 18054–18062 (2016). Article CAS PubMed
Google Scholar * Oren, A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. _Saline Syst._ 4, 2 (2008). Article PubMed PubMed Central CAS Google Scholar
* Padan, E., Venturi, M., Gerchman, Y. & Dover, N. Na+/H+ antiporters. _Biochim. Biophys. Acta_ 1505, 144–157 (2001). Article CAS PubMed Google Scholar * Purvis, J. E., Yomano, L.
P. & Ingram, L. O. Enhanced trehalose production improves growth of _Escherichia coli_ under osmotic stress. _Appl. Environ. Microbiol._ 71, 3761–3769 (2005). Article CAS PubMed
PubMed Central Google Scholar * Yang, L. et al. A primary sodium pump gene of the moderate halophile _Halobacillus dabanensis_ exhibits secondary antiporter properties. _Biochem. Biophys.
Res. Commun._ 346, 612–617 (2006). Article CAS PubMed Google Scholar * Ekberg, J. et al. Adaptive evolution of the lager brewing yeast _Saccharomyces pastorianus_ for improved growth
under hyperosmotic conditions and its influence on fermentation performance. _FEMS Yeast Res._ 13, 335–349 (2013). Article CAS PubMed Google Scholar * Schweikhard, E. S., Kuhlmann, S.
I., Kunte, H. J., Grammann, K. & Ziegler, C. M. Structure and function of the universal stress protein TeaD and its role in regulating the ectoine transporter TeaABC of _Halomonas
elongata_ DSM 2581(T). _Biochemistry_ 49, 2194–2204 (2010). Article CAS PubMed Google Scholar * Kunte, H., Lentzen, G. & Galinski, E. Industrial production of the cell protectant
ectoine: protection mechanisms, processes, and products. _Curr. Biotechnol._ 3, 10–25 (2014). Article CAS Google Scholar * Chen, X., Yu, L., Qiao, G. & Chen, G. Q. Reprogramming
_Halomonas_ for industrial production of chemicals. _J. Ind. Microbiol. Biotechnol._ 45, 545–554 (2018). Article CAS PubMed Google Scholar * Qin, Q. et al. CRISPR/Cas9 editing genome of
extremophile _Halomonas_ spp. _Metab. Eng._ 47, 219–229 (2018). Article CAS PubMed Google Scholar * Chen, R. et al. Optimization of the extraction and purification of the compatible
solute ectoine from _Halomonas_ elongate in the laboratory experiment of a commercial production project. _World J. Microbiol. Biotechnol._ 33, 116 (2017). Article PubMed CAS Google
Scholar * Zaky, A. S., Greetham, D., Tucker, G. A. & Du, C. The establishment of a marine focused biorefinery for bioethanol production using seawater and a novel marine yeast strain.
_Sci. Rep._ 8, 12127 (2018). Article PubMed PubMed Central CAS Google Scholar * Yuan, W. J., Zhao, X. Q., Ge, X. M. & Bai, F. W. Ethanol fermentation with _Kluyveromyces marxianus_
from Jerusalem artichoke grown in salina and irrigated with a mixture of seawater and freshwater. _J. Appl. Microbiol._ 105, 2076–2083 (2008). Article CAS PubMed Google Scholar * Ramos,
J. L., Duque, E., Godoy, P. & Segura, A. Efflux pumps involved in toluene tolerance in _Pseudomonas putida_ DOT-T1E. _J. Bacteriol._ 180, 3323–3329 (1998). Article CAS PubMed PubMed
Central Google Scholar * Rühl, J., Schmid, A. & Blank, L. M. Selected _Pseudomonas putida_ strains able to grow in the presence of high butanol concentrations. _Appl. Environ.
Microbiol._ 75, 4653–4656 (2009). Article PubMed PubMed Central CAS Google Scholar * Ramos, J. L. et al. Mechanisms for solvent tolerance in bacteria. _J. Biol. Chem._ 272, 3887–3890
(1997). Article CAS PubMed Google Scholar * Rojas, A. et al. Three efflux pumps are required to provide efficient tolerance to toluene in _Pseudomonas putida_ DOT-T1E. _J. Bacteriol._
183, 3967–3973 (2001). Article CAS PubMed PubMed Central Google Scholar * Terán, W. et al. Complexity in efflux pump control: cross-regulation by the paralogues TtgV and TtgT. _Mol.
Microbiol._ 66, 1416–1428 (2007). PubMed Google Scholar * Segura, A. et al. Proteomic analysis reveals the participation of energy- and stress-related proteins in the response of
_Pseudomonas putida_ DOT-T1E to toluene. _J. Bacteriol._ 187, 5937–5945 (2005). Article CAS PubMed PubMed Central Google Scholar * Dunlop, M. J. et al. Engineering microbial biofuel
tolerance and export using efflux pumps. _Mol. Syst. Biol._ 7, 487 (2011). Article PubMed PubMed Central Google Scholar * Tan, Z., Yoon, J. M., Nielsen, D. R., Shanks, J. V. &
Jarboe, L. R. Membrane engineering via trans unsaturated fatty acids production improves _Escherichia coli_ robustness and production of biorenewables. _Metab. Eng._ 35, 105–113 (2016).
Article CAS PubMed Google Scholar * Zingaro, K. A. & Terry Papoutsakis, E. GroESL overexpression imparts _Escherichia coli_ tolerance to i-, _n_-, and 2-butanol, 1,2,4-butanetriol
and ethanol with complex and unpredictable patterns. _Metab. Eng._ 15, 196–205 (2013). Article CAS PubMed Google Scholar * Cook, T. B. et al. Genetic tools for reliable gene expression
and recombineering in _Pseudomonas putida_. _J. Ind. Microbiol. Biotechnol._ 45, 517–527 (2018). Article CAS PubMed PubMed Central Google Scholar * Gong, T. et al. Metabolic engineering
of _Pseudomonas putida_ KT2440 for complete mineralization of methyl parathion and γ-hexachlorocyclohexane. _ACS Synth. Biol._ 5, 434–442 (2016). Article CAS PubMed Google Scholar *
Nikel, P. I. & de Lorenzo, V. _Pseudomonas putida_ as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. _Metab. Eng._ 50, 142–155 (2018).
Article CAS PubMed Google Scholar * Bormann, S. et al. Engineering _Clostridium acetobutylicum_ for production of kerosene and diesel blendstock precursors. _Metab. Eng._ 25, 124–130
(2014). Article CAS PubMed Google Scholar * Anbarasan, P. et al. Integration of chemical catalysis with extractive fermentation to produce fuels. _Nature_ 491, 235–239 (2012). Article
CAS PubMed Google Scholar * Zhang, J., Wu, C. D., Du, G. C. & Chen, J. Enhanced acid tolerance in Lactobacillus casei by adaptive evolution and compared stress response during acid
stress. _Biotechnol. Bioprocess Eng._ 17, 283–289 (2012). Article CAS Google Scholar * Zhou, L. et al. Improvement of d-lactate productivity in recombinant _Escherichia coli_ by coupling
production with growth. _Biotechnol. Lett._ 34, 1123–1130 (2012). Article CAS PubMed Google Scholar * Lee, J. Y., Kang, C. D., Lee, S. H., Park, Y. K. & Cho, K. M. Engineering
cellular redox balance in _Saccharomyces cerevisiae_ for improved production of l-lactic acid. _Biotechnol. Bioeng._ 112, 751–758 (2015). Article CAS PubMed Google Scholar * Yan, D. et
al. Construction of reductive pathway in _Saccharomyces cerevisiae_ for effective succinic acid fermentation at low pH value. _Bioresour. Technol._ 156, 232–239 (2014). Article CAS PubMed
Google Scholar * Kwon, Y. D., Kim, S., Lee, S. Y. & Kim, P. Long-term continuous adaptation of _Escherichia coli_ to high succinate stress and transcriptome analysis of the tolerant
strain. _J. Biosci. Bioeng._ 111, 26–30 (2011). Article CAS PubMed Google Scholar * Wright, J. et al. Batch and continuous culture-based selection strategies for acetic acid tolerance in
xylose-fermenting _Saccharomyces cerevisiae_. _FEMS Yeast Res._ 11, 299–306 (2011). Article CAS PubMed Google Scholar * Sandoval, N. R., Mills, T. Y., Zhang, M. & Gill, R. T.
Elucidating acetate tolerance in _E. coli_ using a genome-wide approach. _Metab. Eng._ 13, 214–224 (2011). Article CAS PubMed Google Scholar * Suominen, P. _et al_. Genetically modified
yeast of the species _Issatchenkia Orientalis_ and closely relates species, and fermentation processes using same (Cargill Inc., 2012). * Valdés, J. et al. _Acidithiobacillus ferrooxidans_
metabolism: from genome sequence to industrial applications. _BMC Genomics_ 9, 597 (2008). Article PubMed PubMed Central CAS Google Scholar * Kernan, T. et al. Engineering the
iron-oxidizing chemolithoautotroph _Acidithiobacillus ferrooxidans_ for biochemical production. _Biotechnol. Bioeng._ 113, 189–197 (2016). Article CAS PubMed Google Scholar * Inaba, Y.,
Banerjee, I., Kernan, T. & Banta, S. Transposase-mediated chromosomal integration of exogenous genes in _Acidithiobacillus ferrooxidans_. _Appl. Environ. Microbiol._ 84, e01381–18
(2018). Article CAS PubMed PubMed Central Google Scholar * Maezato, Y., Johnson, T., McCarthy, S., Dana, K. & Blum, P. Metal resistance and lithoautotrophy in the extreme
thermoacidophile _Metallosphaera sedula_. _J. Bacteriol._ 194, 6856–6863 (2012). Article CAS PubMed PubMed Central Google Scholar * Auernik, K. S., Maezato, Y., Blum, P. H. & Kelly,
R. M. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon _Metallosphaera sedula_ provides insights into bioleaching-associated metabolism. _Appl. Environ.
Microbiol._ 74, 682–692 (2008). Article CAS PubMed Google Scholar * Zeldes, B. M. et al. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels
and industrial chemicals. _Front. Microbiol._ 6, 1209 (2015). Article PubMed PubMed Central Google Scholar * Feist, A. M. et al. A genome-scale metabolic reconstruction for _Escherichia
coli_ K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. _Mol. Syst. Biol._ 3, 121 (2007). Article PubMed PubMed Central CAS Google Scholar * Förster, J., Famili,
I., Fu, P., Palsson, B. O. & Nielsen, J. Genome-scale reconstruction of the _Saccharomyces cerevisiae_ metabolic network. _Genome Res._ 13, 244–253 (2003). Article PubMed PubMed
Central CAS Google Scholar * Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. _Nature_ 460, 894–898 (2009). Article CAS PubMed PubMed
Central Google Scholar * Warner, J. R., Reeder, P. J., Karimpour-Fard, A., Woodruff, L. B. & Gill, R. T. Rapid profiling of a microbial genome using mixtures of barcoded
oligonucleotides. _Nat. Biotechnol._ 28, 856–862 (2010). Article CAS PubMed Google Scholar * Lee, M. E., DeLoache, W. C., Cervantes, B. & Dueber, J. E. A highly characterized yeast
toolkit for modular, multipart assembly. _ACS Synth. Biol._ 4, 975–986 (2015). Article CAS PubMed Google Scholar * Xu, P., Vansiri, A., Bhan, N. & Koffas, M. A. ePathBrick: a
synthetic biology platform for engineering metabolic pathways in _E. coli_. _ACS Synth. Biol._ 1, 256–266 (2012). Article CAS PubMed Google Scholar * Löbs, A. K., Schwartz, C. &
Wheeldon, I. Genome and metabolic engineering in non-conventional yeasts: current advances and applications. _Synth. Syst. Biotechnol._ 2, 198–207 (2017). Article PubMed PubMed Central
Google Scholar * Schwartz, C., Frogue, K., Ramesh, A., Misa, J. & Wheeldon, I. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous
recombination in _Yarrowia lipolytica_. _Biotechnol. Bioeng._ 114, 2896–2906 (2017). Article CAS PubMed Google Scholar * Cao, M., Gao, M., Ploessl, D., Song, C. & Shao, Z.
CRISPR-mediated genome editing and gene repression in _Scheffersomyces stipitis_. _Biotechnol. J._ 13, e1700598 (2018). Article PubMed CAS Google Scholar * Schwartz, C., Shabbir-Hussain,
M., Frogue, K., Blenner, M. & Wheeldon, I. Standardized markerless gene integration for pathway engineering in _Yarrowia lipolytica_. _ACS Synth. Biol._ 6, 402–409 (2017). Article CAS
PubMed Google Scholar * Schwartz, C. et al. Validating genome-wide CRISPR-Cas9 function improves screening in the oleaginous yeast _Yarrowia lipolytica_. _Metab. Eng._ 55, 102–110
(2019). Article CAS PubMed Google Scholar * Schwartz, C. M., Hussain, M. S., Blenner, M. & Wheeldon, I. Synthetic RNA polymerase III promoters facilitate high-efficiency
CRISPR-Cas9-mediated genome editing in _Yarrowia lipolytica_. _ACS Synth. Biol._ 5, 356–359 (2016). Article CAS PubMed Google Scholar * Cao, M. et al. Centromeric DNA facilitates
nonconventional yeast genetic engineering. _ACS Synth. Biol._ 6, 1545–1553 (2017). Article CAS PubMed Google Scholar * Lewis, N. E., Nagarajan, H. & Palsson, B. O. Constraining the
metabolic genotype-phenotype relationship using a phylogeny of in silico methods. _Nat. Rev. Microbiol._ 10, 291–305 (2012). Article CAS PubMed PubMed Central Google Scholar * Schwalen,
C. J., Hudson, G. A., Kille, B. & Mitchell, D. A. Bioinformatic expansion and discovery of thiopeptide antibiotics. _J. Am. Chem. Soc._ 140, 9494–9501 (2018). Article CAS PubMed
PubMed Central Google Scholar * Brophy, J. A. N. et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. _Nat.
Microbiol._ 3, 1043–1053 (2018). THIS WORK ENGINEERS THE INTEGRATIVE AND CONJUGATIVE ELEMENTS OF _B__ACILLUS SUBTILIS_ TO CREATE A STRAIN TECHNOLOGY CALLED XPORT THAT CAN BE USED TO RAPIDLY
ENGINEER NOVEL BACTERIAL ISOLATES. Article CAS PubMed Google Scholar * Schwartz, C., Curtis, N., Löbs, A. K. & Wheeldon, I. Multiplexed CRISPR activation of cryptic sugar metabolism
enables _Yarrowia lipolytica_ growth on cellobiose. _Biotechnol. J._ 13, e1700584 (2018). Article PubMed CAS Google Scholar * Boundy-Mills, K. L. et al. Yeast culture collections in the
twenty-first century: new opportunities and challenges. _Yeast_ 33, 243–260 (2016). Article CAS PubMed Google Scholar * Solomon, K. V. et al. Early-branching gut fungi possess a large,
comprehensive array of biomass-degrading enzymes. _Science_ 351, 1192–1195 (2016). THIS WORK USES ‘OMICS’-LEVEL ANALYSES AND BIOCHEMICAL ASSAYS TO DISCOVER NEW BIOTECHNOLOGY-RELEVANT
ENZYMES, INCLUDING CELLULASES FOR THE BREAKDOWN OF LIGNOCELLULOSIC BIOMASS IN NEW ISOLATES OF ANAEROBIC GUT FUNGI. Article CAS PubMed PubMed Central Google Scholar Download references
ACKNOWLEDGEMENTS This material is based upon work supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program under
Award Number DE-SC0019093, Air Force Office of Scientific Research award FA9550-17-1-0270, Army Research Office MURI award W911NF1410263, and National Science Foundation award NSF-CBET
1706545 for funding. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Chemical and Environmental Engineering, University of California Riverside, Riverside, CA, USA Sarah
Thorwall, Cory Schwartz & Ian Wheeldon * Department of Bioengineering, University of California Riverside, Riverside, CA, USA Justin W. Chartron * Center for Industrial Biotechnology,
Bourns College of Engineering, University of California Riverside, Riverside, CA, USA Ian Wheeldon Authors * Sarah Thorwall View author publications You can also search for this author
inPubMed Google Scholar * Cory Schwartz View author publications You can also search for this author inPubMed Google Scholar * Justin W. Chartron View author publications You can also search
for this author inPubMed Google Scholar * Ian Wheeldon View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Ian
Wheeldon. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Thorwall, S., Schwartz, C.,
Chartron, J.W. _et al._ Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. _Nat Chem Biol_ 16, 113–121 (2020). https://doi.org/10.1038/s41589-019-0452-x
Download citation * Received: 06 February 2019 * Accepted: 11 December 2019 * Published: 23 January 2020 * Issue Date: February 2020 * DOI: https://doi.org/10.1038/s41589-019-0452-x SHARE
THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Real madrid will call tottenham boss mauricio pochettino - balagueZidane resigned as head coach of Real Madrid just days after leading the Spanish club to a third straight Champions Leag...
Were team gb’s skeleton suits responsible for fantastic three medal haul?Team GB skeleton rider Lizzie Yarnold won a stunning Winter Olympic gold on February 17, backed up by bronzes for Laura ...
9/11 first responder pays it forward with relief missionsMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...
Certificates programs can boost your career and skillsDerrick Lewis has new appreciation for something a coworker told him 20 years ago: “You have to keep learning.” That adv...
Comment: weeds can make a garden cheaper and more manageable in franceCOLUMNIST SAMANTHA DAVID TELLS HOW EMBRACING NATURE HAS HIDDEN BENEFITS The day after I moved into my French farmhouse, ...
Latests News
Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesisABSTRACT Microbial chemical production is a rapidly growing industry, with much of the growth fueled by advances in synt...
Commentary: millennials are $1 trillion in debt, but they're better at savingNew findings from the New York Federal Reserve reveal that millennials have now racked up over $1 trillion of debt. This...
Linking lysosome function to macrophage homeostasisAccess through your institution Buy or subscribe A recent study in _Science_ reveals that equilibrative nucleoside trans...
Page not found - ABC KidsSorry, page not found This might be because:The page you were looking for was removed, had its name changed, or is tempo...
Authority might rescind controversial fee : schools: joint panel of trustees will reconsider annual levy in wake of strong opposition. Legal challengeHUNTINGTON BEACH — Less than two weeks after imposing a controversial $50-per-year fee on property owners to maintain sc...