A swapped genetic code prevents viral infections and gene transfer
A swapped genetic code prevents viral infections and gene transfer"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Engineering the genetic code of an organism has been proposed to provide a firewall from natural ecosystems by preventing viral infections and gene transfer1,2,3,4,5,6. However, numerous
viruses and mobile genetic elements encode parts of the translational apparatus7,8,9, potentially rendering a genetic-code-based firewall ineffective. Here we show that such mobile transfer
RNAs (tRNAs) enable gene transfer and allow viral replication in Escherichia coli despite the genome-wide removal of 3 of the 64 codons and the previously essential cognate tRNA and release
factor genes. We then establish a genetic firewall by discovering viral tRNAs that provide exceptionally efficient codon reassignment allowing us to develop cells bearing an amino
acid-swapped genetic code that reassigns two of the six serine codons to leucine during translation. This amino acid-swapped genetic code renders cells resistant to viral infections by
mistranslating viral proteomes and prevents the escape of synthetic genetic information by engineered reliance on serine codons to produce leucine-requiring proteins. As these cells may have
a selective advantage over wild organisms due to virus resistance, we also repurpose a third codon to biocontain this virus-resistant host through dependence on an amino acid not found in
nature10. Our results may provide the basis for a general strategy to make any organism safely resistant to all natural viruses and prevent genetic information flow into and out of
genetically modified organisms.
Raw data from whole-genome sequencing, transcriptome and tRNA-seq experiments have been deposited to the Sequence Read Archive under the BioProject ID PRJNA856259. tRNA data and the
generated sequences in this study are included in the Supplementary Data files. Mass spectra and proteome measurements have been deposited to MassIVE (MSV000089854;
https://doi.org/10.25345/C5FF3M41W)65. The Syn61Δ3(ev5) ΔrecA (ev1) strain is available from Addgene (bacterial strain no. 189857). We cannot deposit Ec_Syn61∆3-SL and Ec_Syn61∆3 adk.d6 at
Addgene owing to the incompatibility of Addgene’s methods of strain distribution and growth medium requirements, but all materials used in this study are freely available from the
corresponding authors for academic research use upon request. The PHROG HMM database is available at https://phrogs.lmge.uca.fr and from ref. 51. The assembled annotated genome of E. coli
Syn61 substrain Syn61Δ3(ev5) is available in Supplementary Data 4, and the annotated genomes of REP phages have been deposited to NCBI GenBank under the accession numbers OQ174500, OQ174501,
OQ174502, OQ174503, OQ174504, OQ174505, OQ174506, OQ174507, OQ174508, OQ174509, OQ174510 and OQ174511. Source data are provided with this paper.
We thank G. Pósfai (Biological Research Centre, Hungary) for sharing MDS42 and J. W. Chin’s team (Medical Research Council Laboratory of Molecular Biology, UK) for sharing Syn61∆3 through
Addgene. Financial support for this research was provided by the US Department of Energy under grant DE-FG02-02ER63445 and by National Science Foundation award number 2123243 (both to
G.M.C.). M.B. acknowledges support from the NIGMS of the National Institutes of Health (R35GM133700), the David and Lucile Packard Foundation, the Pew Charitable Trusts and the Alfred P.
Sloan Foundation. A.N. was supported by the EMBO LTF 160–2019 Long-Term Fellowship. We thank A. Millard’s laboratory for making the PHROG HMM database available for bacteriophage annotation,
GenScript USA Inc. for their DNA synthesis support, and D. Snyder, K. Harris and all members of the MiGS, Pittsburgh, PA for their support with DNA and RNA sequencing. We are grateful to T.
Wu for support, Y. Shen and S. Yan (Institute of Biochemistry, Beijing Genomics Institute) for collaboration on genome recoding, and B. Hajian for graphical design and help with
illustrations.
Present address: Department of Chemical Engineering, University of Washington, Seattle, WA, USA
Department of Genetics, Harvard Medical School, Boston, MA, USA
Akos Nyerges, Svenja Vinke, Regan Flynn, Kamesh Narasimhan, Jorge A. Marchand, Maximilien Baas-Thomas, Anush Chiappino-Pepe & George M. Church
Department of Biomedical Informatics and Laboratory of Systems Pharmacology, Harvard Medical School, Boston, MA, USA
Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA, USA
Department of Pathology and Immunology, Washington University School of Medicine in St. Louis, St Louis, MO, USA
The Edison Family Center for Genome Sciences and Systems Biology, Washington University School of Medicine in St. Louis, St Louis, MO, USA
A.N. developed the project, led analyses and wrote the manuscript with input from all authors. A.N. and G.M.C. supervised research. S.V. carried out tRNALeu suppressor screens and sfGFP
expression assays, and assisted in the construction of the pLS plasmids and biocontainment experiments. R.F. assisted in experiments, and carried out adaptive laboratory evolution and growth
rate measurements. S.V.O., E.A.R. and M.B. provided environmental samples for phage isolation, carried out replication assays and provided support for phage experiments and genome analyses.
M.L., K.C. and F.H. carried out DNA synthesis; M.B.-T., A.C.-P. and E.K. supported the project. B.B. carried out MS/MS analyses. K.N. and J.A.M. provided reagents for tRNA-seq experiments.
Harvard Medical School has filed a provisional patent application related to this work on which A.N., S.V. and G.M.C. are listed as inventors. M.L., K.C. and F.H. are employed by GenScript
USA Inc., but the company had no role in designing or executing experiments. G.M.C. is a founder of the following companies in which he has related financial interests: GRO Biosciences,
EnEvolv (Ginkgo Bioworks) and 64x Bio. Other potentially relevant financial interests of G.M.C. are listed at http://arep.med.harvard.edu/gmc/tech.html.
Nature thanks Benjamin Blount and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
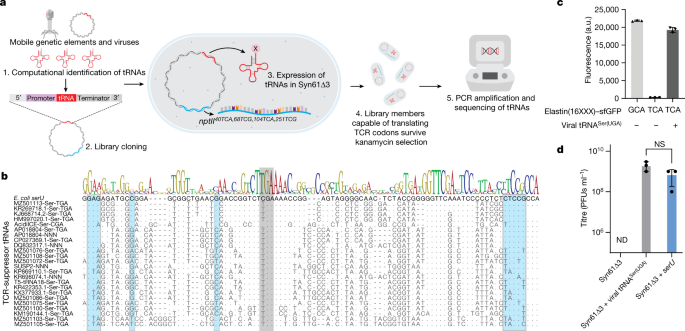
a) Viral TCR suppressor tRNAs decode TCA codons as serine. The amino acid identity of the translated TCR codon within elastin16 TCA-sfGFP-His6 was confirmed by tandem mass spectrometry from
Syn61∆3 expressing the tRNA-SerUGA of Escherichia phage IrisVonRoten (GenBank ID MZ501075)24. The figure shows the amino acid sequence and MS/MS spectrum of the analyzed elastin16 TCA
peptide. MS/MS data was collected once. b) Syn61∆3 obstructs the replication of viruses containing genomic tRNA-SerUGA. Figure shows the titer of twelve tRNA gene-containing coliphages24,
after 24 h of growth on MDS42 and Syn61∆3. All analyzed bacteriophages, except MZ501058, contain a genomic tRNA-SerUGA tRNA that provides TCR suppressor activity based on our screen (Fig.
1b, Supplementary Data 1). Early exponential phase cultures of MDS42 and Syn61∆3 were infected at an MOI of 0.001 with the corresponding phages, and free phage titers were determined after
24 h of incubation. Measurements were performed in n = 3 independent experiments (i.e., MDS42 + MZ501058, MZ501065, MZ501075, MZ501105, MZ501106; Syn61∆3 + MZ501046, MZ501067, MZ501066,
MZ501096, MZ501074, MZ501098) or in n = 2 independent experiments (i.e., MDS42 + MZ501046, MZ501066, MZ501067, MZ501074, MZ501089, MZ501096, MZ501098; Syn61∆3 + MZ501058, MZ501065, MZ501075,
MZ501089, MZ501105, MZ501106); dashed line represents input phage titer without bacterial cells (i.e., a titer of 420 PFU/ml); dots represent data from independent experiments; bar graphs
represent the mean; error bars represent the SEM based on n = 3 independent experiments.
The time-course kinetics of host and viral tRNA expression in Syn61∆3 cells following REP12 phage infection was quantified using tRNAseq (Methods). The endogenous serV and serW tRNAs of the
host Syn61∆3 are highlighted in green and blue, respectively, while the tRNA-SerUGA of the REP12 virus is highlighted in red. REP12 viral tRNAs are shown in light red; endogenous tRNAs of
Syn61∆3 are shown in gray. Data represent mean TPM (transcript/million). Source data is available within this paper.
The tRNA tail is modified into CCA even if the phage-encoded tRNA-SerUGA does not encode a CCA end. The sequence of genomic phage-encoded tRNA-SerUGA is highlighted in magenta. Black letters
indicate example tRNAseq sequencing reads from REP12 infected Syn61∆3 cells directly after phage attachment based on a single experiment.
Volcano plot shows the differential expression between uninfected and REP12-infected Syn61∆3 cells, 40 min post-infection, based on n = 3 independent experiments (Methods). QueG encodes
epoxyqueuosine reductase that catalyzes the final step in the de novo synthesis of queuosine in tRNAs. TrmJ encodes tRNA Cm32/Um32 methyltransferase that introduces methyl groups at the 2′-O
position of U32 of several tRNAs, including tRNA-SerUGA. Differential expression and -Log10 adjusted p-values were calculated using the DESeq2 algorithm55. Source data is available within
this paper.
a) Multiple sequence alignment of leuUYGA variants selected in aminoglycoside O-phosphotransferase expression screen, compared to E. coli leuU and the YGA anticodon-swapped E. coli leuU tRNA
variant. Grey shading indicates the anticodon region, and the host’s LeuS leucine-tRNA-ligase identity elements31 are shown in blue. Sequence information of the leuUYGA variants is
available in Supplementary Data 3. b) Multiple sequence alignment of phage-derived tRNA-LeuYGA variants selected in the aph3Ia29×Leu→TCR aminoglycoside O-phosphotransferase expression
screen, compared to endogenous E. coli leucine tRNAs. Grey shading indicates the anticodon region, while the host’s LeuS leucine-tRNA-ligase identity elements31 are shown in blue. Sequence
information of the phage-derived tRNA-LeuYGA variants is available in Supplementary Data 3.
tRNA levels of Ec_Syn61∆3-SL were quantified using tRNAseq (Methods). Viral Leu-tRNAUGA and Leu-tRNACGA are highlighted in red, and the host’s endogenous E. coli serV tRNA is highlighted in
orange. tRNAseq data was collected once. Data represent TPM (transcript/million). Source data is available within this paper.
a) MS/MS spectrum of the serine-to-leucine mistranslated TufA peptide. b) MS/MS spectrum of the wild-type TufA peptide. The figure shows the amino acid sequence and MS/MS spectrum of the
Ec_Syn61∆3-SL-expressed TufA protein fragment, together with its genomic sequence, in which the serine-coding TCT codon (as shown in panel b) is partially mistranslated as leucine (as shown
in panel a). The experiment was performed by analyzing the total proteome of Ec_Syn61∆3-SL cells, expressing Leu9-tRNAYGA from Escherichia phage OSYSP (GenBank ID MF402939.1) by tandem mass
spectrometry (Methods). MS/MS data was collected once.
a) Doubling times of Syn61Δ3(ev5), Syn61Δ3(ev5)ΔrecA, Syn61Δ3(ev5)ΔrecA(ev1), and Ec_Syn61∆3-SL, calculated based on growth curves (shown in panels b, c, d) in rich bacterial media under
standard laboratory conditions. b) Growth curves of Syn61Δ3(ev5), Syn61Δ3(ev5)ΔrecA, and Syn61Δ3(ev5)ΔrecA(ev1) in LBL broth. c) Growth curves of Syn61Δ3(ev5) and Syn61Δ3(ev5)ΔrecA(ev1) in
2×YT broth. d) Growth curves of Ec_Syn61∆3-SL in 2×YT broth containing 50 μg/ml kanamycin. Three independent cultures were grown aerobically in vented shake flasks at 37 °C, and OD600
measurements were taken during exponential growth (Methods). Data curves and bars represent the mean. Error bars show standard deviation based on n = 3 independent experiments.
(a) Phage enrichment experiment using Ec_Syn61∆3-SL, expressing KP869110.1 tRNA24 LeuYGA and MF402939.1 tRNA9 LeuYGA, as host. (b) Phage enrichment experiment using Ec_Syn61∆3-SL, expressing
KP869110.1 tRNA24 LeuYGA and MF402939.1 tRNA21 LeuYGA, as host. Phage enrichment experiments were performed by mixing early exponential cultures of Ec_Syn61∆3-SL with 10 ml environmental
sample mix containing the mixture of Sample 2–13 from our study (Extended Data Table 1a). After two enrichment cycles (Methods, Supplementary Note), filter-sterilized culture supernatants
were mixed with phage-susceptible E. coli MDS42 cells in top agar and plated on LBL agar plates to determine viral titer. Enrichment experiments were performed in n = 2 independent
replicates with the same result. (c) Lytic E. coli MDS42 phage plaques after 103-fold dilution of the environmental sample mix. d) Lytic phage titer of the environmental sample mix, before
and after enrichment on Ec_Syn61∆3-SL. Dots represent the viral titer of the unenriched sample based on three independent experiments, measured on E. coli MDS42 cells. ND represents no
plaques detected. Bar represents the mean. Error bar shows standard deviation based on n = 3 independent experiments.
This file contains Supplementary Note, Figs. 1–3 and references.
tRNA candidates within tRNA library for TCR suppressor experiments; Identified TCR suppressor tRNAs; All predicted tRNAs within NCBI RefSeq Prokaryotic viral genomes; All predicted tRNAs
within REP1–REP12 phage isolates.
DNA constructs and primers; tRNA libraries; Mutations of Syn61Δ3(ev5) ΔrecA (ev1); Strains and plasmids used in this study.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author
self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Anyone you share the following link with will be able to read this content:
Trending News
404 Error - Los Angeles TimesSorry! The page you were looking for cannot be found.The page may no longer exist or may have moved to another web addre...
The 9 worst habits for your brain4. YOU HAVE UNHEALTHY SLEEP HABITS Quality sleep is crucial to a sharp and productive mind, according to the Global Coun...
Dual Ni/Co-hemin metal–organic framework-PrGO for high-performance asymmetric hybrid supercapacitorDownload PDF Article Open access Published: 01 August 2023 Dual Ni/Co-hemin metal–organic framework-PrGO for high-perfor...
Old state pension pays £45 a week LESS than new one from April - ‘we're being robbed'Those who retired before April 6, 2016, on the old basic State Pension are furious at falling behind. However, pension e...
Secure .NET CORE 6 Web API with JWT from Duende IdentityServerMember-only storySecure .NET CORE 6 Web API with JWT from Duende IdentityServerFuji NguyenFollow6 min read·Jun 2, 2022 -...
Latests News
A swapped genetic code prevents viral infections and gene transferEngineering the genetic code of an organism has been proposed to provide a firewall from natural ecosystems by preventin...
Chinese cities pull global luxury property prices higher as taxes bite elsewhereGrowth in luxury house prices of 36.2 percent in China's third largest city, Guangzhou, has helped to bump the global in...
How to watch the ny knicks in the 2024 nba playoffs: tv channels, streamingNew York Post may be compensated and/or receive an affiliate commission if you click or buy through our links. Featured ...
Jean-Georges and the Opening of Perry St - A Return to the Clean Cuisine He Made Famous - NymagJean-Georges is Seeing Stars By Jay McInerney June 17, 2005 saved Save this article to read it later. Find this story in...
Unexpectedly strong hydrophilic character of free-standing thin films from carbon nanotubesABSTRACT We report on the development of a method of formation of hydrophilic carbon nanotube (CNT) films. The technique...