Machine learning-aided engineering of hydrolases for pet depolymerization
Machine learning-aided engineering of hydrolases for pet depolymerization"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Plastic waste poses an ecological challenge1,2,3 and enzymatic degradation offers one, potentially green and scalable, route for polyesters waste recycling4. Poly(ethylene
terephthalate) (PET) accounts for 12% of global solid waste5, and a circular carbon economy for PET is theoretically attainable through rapid enzymatic depolymerization followed by
repolymerization or conversion/valorization into other products6,7,8,9,10. Application of PET hydrolases, however, has been hampered by their lack of robustness to pH and temperature ranges,
slow reaction rates and inability to directly use untreated postconsumer plastics11. Here, we use a structure-based, machine learning algorithm to engineer a robust and active PET
hydrolase. Our mutant and scaffold combination (FAST-PETase: functional, active, stable and tolerant PETase) contains five mutations compared to wild-type PETase (N233K/R224Q/S121E from
prediction and D186H/R280A from scaffold) and shows superior PET-hydrolytic activity relative to both wild-type and engineered alternatives12 between 30 and 50 °C and a range of pH levels.
We demonstrate that untreated, postconsumer-PET from 51 different thermoformed products can all be almost completely degraded by FAST-PETase in 1 week. FAST-PETase can also depolymerize
untreated, amorphous portions of a commercial water bottle and an entire thermally pretreated water bottle at 50 ºC. Finally, we demonstrate a closed-loop PET recycling process by using
FAST-PETase and resynthesizing PET from the recovered monomers. Collectively, our results demonstrate a viable route for enzymatic plastic recycling at the industrial scale. Access through
your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature
Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 51 print issues and online access
$199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are
calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
POLYESTER-DEGRADING ENZYMES IN A CIRCULAR ECONOMY OF PLASTICS Article 12 May 2025 DISCOVERY AND MECHANISM-GUIDED ENGINEERING OF BHET HYDROLASES FOR IMPROVED PET RECYCLING AND UPCYCLING
Article Open access 13 July 2023 PROCESS INNOVATIONS TO ENABLE VIABLE ENZYMATIC POLY(ETHYLENE TEREPHTHALATE) RECYCLING Article 06 May 2025 DATA AVAILABILITY The authors declare that all data
supporting the findings of this study are available in the article, its Extended Data, its Source Data or from the corresponding authors upon request. The complete data set of MutCompute
predictions used in this study can be acquired at https://mutcompute.com. Coordinates for the FAST-PETase structure have been deposited into the PDB with accession code 7SH6. Interactive
visualizations of MutCompute for Fig. 1 are available at https://www.mutcompute.com/petase/5xjh and https://www.mutcompute.com/petase/6ij6. Source data are provided with this paper. CODE
AVAILABILITY MutCompute and MutCompute-View are publicly available at https://mutcompute.com and https://mutcompute.com/view for academic research. REFERENCES * Geyer, R., Jambeck, J. R.
& Law, K. L. Production, use, and fate of all plastics ever made. _Sci. Adv._ 3, e1700782 (2017). Article ADS Google Scholar * Santos, R. G., Machovsky-Capuska, G. E. & Andrades,
R. Plastic ingestion as an evolutionary trap: toward a holistic understanding. _Science_ 373, 56–60 (2021). Article CAS ADS Google Scholar * MacLeod, M., Arp, H. P. H., Tekman, M. B.
& Jahnke, A. The global threat from plastic pollution. _Science_ 373, 61–65 (2021). Article CAS ADS Google Scholar * Chen, C. C., Dai, L., Ma, L. & Guo, R. T. Enzymatic
degradation of plant biomass and synthetic polymers. _Nat. Rev. Chem._ 4, 114–126 (2020). * George, N. & Kurian, T. Recent developments in the chemical recycling of postconsumer
poly(ethylene terephthalate) waste. _Ind. Eng. Chem. Res._ 53, 14185–14198 (2014). Article CAS Google Scholar * Simon, N. et al. A binding global agreement to address the life cycle of
plastics. _Science_ 373, 43–47 (2021). Article CAS ADS Google Scholar * Kawai, F., Kawabata, T. & Oda, M. Current knowledge on enzymatic PET degradation and its possible application
to waste stream management and other fields. _Appl. Microbiol. Biotechnol._ 103, 4253–4268 (2019). Article CAS Google Scholar * Sarah, K. & Gloria, R. Achieving a circular bioeconomy
for plastics. _Science_ 373, 49–50 (2021). Article Google Scholar * Ru, J., Huo, Y. & Yang, Y. Microbial degradation and valorization of plastic wastes. _Front. Microbiol._
https://doi.org/10.3389/fmicb.2020.00442 (2020). * Ellis, L. D. et al. Chemical and biological catalysis for plastics recycling and upcycling. _Nat. Catal._ 4, 539–556 (2021). Article CAS
Google Scholar * Taniguchi, I. et al. Biodegradation of PET: current status and application aspects. _ACS Catal._ https://doi.org/10.1021/acscatal.8b05171 (2019). * Tournier, V. et al. An
engineered PET depolymerase to break down and recycle plastic bottles. _Nature_ 580, 216–219 (2020). Article CAS ADS Google Scholar * Inderthal, H., Tai, S. L. & Harrison, S. T. L.
Non-hydrolyzable plastics – an interdisciplinary look at plastic bio-oxidation. _Trends Biotechnol._ 39, 12–23 (2021). Article CAS Google Scholar * Yoshida, S. et al. A bacterium that
degrades and assimilates poly(ethylene terephthalate). _Science_ 351, 1196–1199 (2016). Article CAS ADS Google Scholar * Chen, C. C. et al. General features to enhance enzymatic activity
of poly(ethylene terephthalate) hydrolysis. _Nat. Catal._ https://doi.org/10.1038/s41929-021-00616-y (2021). * Worm, B., Lotze, H. K., Jubinville, I., Wilcox, C. & Jambeck, J. Plastic
as a persistent marine pollutant. _Ann. Rev. Env. Resourc._https://doi.org/10.1146/annurev-environ-102016-060700 (2017). * Son, H. F. et al. Rational protein engineering of thermo-stable
PETase from _Ideonella sakaiensis_ for highly efficient PET degradation. _ACS Catal._ 9, 3519–3526 (2019). Article CAS Google Scholar * Austin, H. P. et al. Characterization and
engineering of a plastic-degrading aromatic polyesterase. _Proc. Natl Acad. Sci. USA_ 115, E4350–E4357 (2018). Article CAS Google Scholar * Joo, S. et al. Structural insight into
molecular mechanism of poly(ethylene terephthalate) degradation. _Nat. Commun._ 9, 382 (2018). Article ADS Google Scholar * Han, X. et al. Structural insight into catalytic mechanism of
PET hydrolase. _Nat. Commun._ 8, 2106 (2017). Article ADS Google Scholar * Furukawa, M., Kawakami, N., Oda, K. & Miyamoto, K. Acceleration of enzymatic degradation of poly(ethylene
terephthalate) by surface coating with anionic surfactants. _Chem. Sus. Chem._ 11, 4018–4025 (2018). Article CAS Google Scholar * Cui, Y. et al. Computational redesign of a PETase for
plastic biodegradation under ambient condition by the GRAPE strategy. _ACS Catal._ https://doi.org/10.1021/acscatal.0c05126 (2021). * Chen, K., Hu, Y., Dong, X. & Sun, Y. Molecular
insights into the enhanced performance of ekylated petase toward PET degradation. _ACS Catal._ 11, 7358–7370 (2021). Article CAS Google Scholar * Shroff, R. et al. Discovery of novel
gain-of-function mutations guided by structure-based deep learning. _ACS Synth. Biol._ 9, 2927–2935 (2020). Article CAS Google Scholar * Kawai, F. et al. A novel Ca2+-activated,
thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from _Saccharomonospora viridis_ AHK190. _Appl. Microbiol. Biotechnol._ 98, 10053–10064 (2014). Article CAS
Google Scholar * Weissmann, D. Applied Plastics Engineering Handbook: Processing, Materials, and Applications 2nd edn (ed. Kutz, M.) 717–741 (William Andrew Publishing, 2017). * Wallace, N.
E. et al. The highly crystalline PET found in plastic water bottles does not support the growth of the PETase-producing bacterium _Ideonella sakaiensis_. _Environ. Microbiol. Rep._ 12,
578–582 (2020). Article CAS Google Scholar * Wei, R. & Zimmermann, W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: how far are we? _Microb.
Biotechnol._ 10, 1308–1322 (2017). Article CAS Google Scholar * Kawai, F., Kawabata, T. & Oda, M. Current state and perspectives related to the polyethylene terephthalate hydrolases
available for biorecycling. _ACS Sustain. Chem. Eng._ 8, 8894–8908 (2020). Article CAS Google Scholar * Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in
oscillation mode. _Methods Enzymol._ 276, 307–326 (1997). Article CAS Google Scholar * Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. _Acta Crystallogr. D.
Biol. Crystallogr._ 60, 2126–2132 (2004). Article Google Scholar * Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments
in Phenix. _Acta Crystallogr. Sect. D, Struct. Biol._ 75, 861–877 (2019). Article CAS Google Scholar * Fujita, M. et al. Cloning and nucleotide sequence of the gene (amyP) for
maltotetraose-forming amylase from _Pseudomonas stutzeri_ MO-19. _J. Bacteriol._ 171, 1333–1339 (1989). Article CAS Google Scholar * Leonard, S. P. et al. Genetic engineering of bee gut
microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. _ACS Synth. Biol._ 7, 1279–1290 (2018). Article CAS Google Scholar Download references
ACKNOWLEDGEMENTS This work was financed under research agreement no. EM10480.26/UTA16-000509 between the ExxonMobil Research and Engineering Company and The University of Texas at Austin.
Sequencing was conducted at the Genomic Sequencing and Analysis Facility (RRID no. SCR_021713), SEM was conducted at the Microscopy and Imaging Facility (RRID no. SCR_021756) at the UT
Austin Center for Biomedical Research Support, and AFM analysis was conducted at the Texas Materials Institute at UT Austin. N.A.L. and C.Z. thank the Welch Foundation for partial support of
this research (Grant #F-1904). The crystallography study is supported by a grant from the National Institutes of Health (no. GM104896 to Y.J.Z.). Crystallographic data collections were
conducted at Advanced Photon Sources (BL23-ID-B), Department of Energy national user facility. We acknowledge the Texas Advanced Computing Center at The University of Texas at Austin for
providing deep learning resources for neural network predictions and analysis that have contributed to the research results reported in this paper. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * McKetta Department of Chemical Engineering, The University of Texas at Austin, Austin, TX, USA Hongyuan Lu, Natalie J. Czarnecki, Congzhi Zhu, Wantae Kim, Hannah O. Cole,
Nathaniel A. Lynd & Hal S. Alper * Department of Chemistry, The University of Texas at Austin, Austin, TX, USA Daniel J. Diaz * Department of Molecular Biosciences, The University of
Texas at Austin, Austin, TX, USA Raghav Shroff, Daniel J. Acosta, Bradley R. Alexander, Hannah O. Cole, Yan Zhang & Andrew D. Ellington * DEVCOM ARL-South, Austin, TX, USA Raghav Shroff
Authors * Hongyuan Lu View author publications You can also search for this author inPubMed Google Scholar * Daniel J. Diaz View author publications You can also search for this author
inPubMed Google Scholar * Natalie J. Czarnecki View author publications You can also search for this author inPubMed Google Scholar * Congzhi Zhu View author publications You can also search
for this author inPubMed Google Scholar * Wantae Kim View author publications You can also search for this author inPubMed Google Scholar * Raghav Shroff View author publications You can
also search for this author inPubMed Google Scholar * Daniel J. Acosta View author publications You can also search for this author inPubMed Google Scholar * Bradley R. Alexander View author
publications You can also search for this author inPubMed Google Scholar * Hannah O. Cole View author publications You can also search for this author inPubMed Google Scholar * Yan Zhang
View author publications You can also search for this author inPubMed Google Scholar * Nathaniel A. Lynd View author publications You can also search for this author inPubMed Google Scholar
* Andrew D. Ellington View author publications You can also search for this author inPubMed Google Scholar * Hal S. Alper View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS H.S.A., A.D.E., N.A.L. and H.L. designed and directed the research. In investigation and validation, R.S. and D.J.D. performed neural network analysis.
H.L. performed enzyme engineering, purification and the depolymerization experiments of both model and pc-PET substrates. H.L., N.J.C., C.Z., D.J.A. and H.O.C. carried out structural and
physical characterization of variants. H.L., C.Z. and N.J.C. performed physical characterization of the treated and untreated commercial PET materials. C.Z carried out experiments for
purifying TPA and regenerating virgin PET and plastics films. D.J.D. and B.R.A. developed MutCompute-View for visualizing predictions from the neural network model. W.K. and Y.J.Z. performed
protein crystallization and structural analysis of the engineered enzyme. H.S.A. and H.L. wrote the original draft of the manuscript. H.S.A., A.D.E., N.A.L. and H.L. revised the manuscript.
H.S.A. and A.D.E. conceived the project idea. All authors reviewed and accepted the manuscript. CORRESPONDING AUTHOR Correspondence to Hal S. Alper. ETHICS DECLARATIONS COMPETING INTERESTS
A patent has been filed in 2020, ‘Mutations for improving activity and thermostability of PETase enzymes’ relating to the mutants and applications developed in this study. R.S. is a
cofounder of Aperiam, a company that applies machine learning to protein engineering. R.S. and A.D.E. are inventors on a patent for applying machine learning to protein engineering that has
been licensed to Aperiam. PEER REVIEW PEER REVIEW INFORMATION _Nature_ thanks Gregory Bowman, Ulphard Thoden van Velzen and the other, anonymous, reviewer(s) for their contribution to the
peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
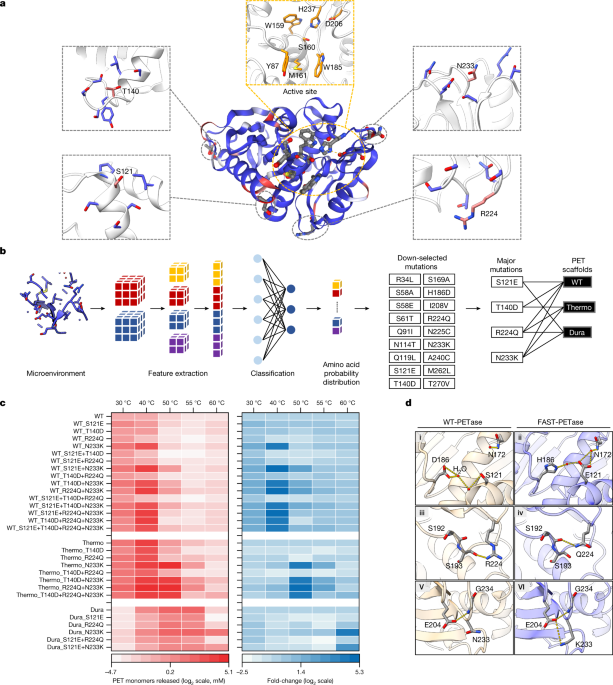
EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIG. 1 TOP 10 MUTCOMPUTE PREDICTIONS RANKED BY FOLD CHANGE IN THE PROBABILITIES BETWEEN THE PREDICTED AND THE WILD-TYPE AMINO ACID. The top 10
mutations predicted using the wild-type PETase (A) and ThermoPETase (B) as scaffolds are presented. MutCompute is an ensembled model that consist of three individually trained 3-dimensional
convolutional neural network (3DCNN) models. Thus, the avg_log_ratio column is the average of the three log ratio values obtained from the three 3DCNN models, rather than being the log ratio
of the average probability assigned to the wild type and predicted amino acid across the three 3DCNN models. EXTENDED DATA FIG. 2 THERMOSTABILITY AND PROTEIN YIELD OF THE PETASE VARIANTS
INCORPORATING THE PREDICTED MUTATIONS AND THEIR RESPECTIVE SCAFFOLDS—WILD-TYPE PETASE (WT), THERMOPETASE (THERMO), DURAPETASE (DURA). Tm of each enzyme (left) was determined by DSC. The
protein yield of each enzyme (right) from _P. putida_ purification experiments was evaluated using a Bradford protein assay. All measurements were conducted in triplicate (n = 3). The bars
shown represent the average numbers. EXTENDED DATA FIG. 3 X-RAY CRYSTAL STRUCTURE OF FAST-PETASE. A, Overall crystal structure of FAST-PETase. Catalytic triads (S160, D206, H237) are shown
in blue sticks. Mutations originating from or shared with ThermoPETase (S121E, D186H, R280A) are shown in pink sticks, and completely novel mutations predicted by MutCompute are shown in
green-yellow sticks. B, C, 2Fo-Fc map (contoured at 1.5 σ) shown as grey mesh superimposed on the stick models of novel mutation sites (B) R224Q, (C) N233K. EXTENDED DATA FIG. 4 LOCATION OF
THE LM SITE IN THE CRYSTAL STRUCTURES OF HOMOLOGOUS PHES. On wild-type PETase from _I. Sakaiensis_ (white ribbon), catalytic residues are shown as blue sticks. LM site is shown as gray
sticks on top of cartoon representation. LM site is zoomed in to show superimposed structures of four different homologous PHEs (right top panel: WT): _I. Sakaiensis_ wild-type PETase (gray
sticks, PDB code 5XJH), LCC (yellow sticks, PDB code 4EB0), LCCF243I/D238C/S283C/N246M (ICCM) (green sticks, PDB code 6THT - LCCF243I/D238C/S283C/Y127G (ICCG) variant structure), and _S.
viridis_ Cut190 (pink sticks, PDB code 4WFI - Cut190S226P variant structure). Based on FAST-PETase structure, the structure of homologous PHEs with LM is modelled (right bottom panel: LM)
with residues shown as blue-colored sticks. EXTENDED DATA FIG. 5 TIME-COURSE OF MASS LOSS AND PET MONOMERS RELEASED FROM HYDROLYZING THE HOLE-PUNCHED FILMS OF SIX REPRESENTATIVE PC-PET
PRODUCTS WITH FAST-PETASE. The six pc-PET products represent PET #2, 6, 8, 25, 29, 32 that were randomly selected from the 51 pc-PET products (Supplementary Table 3 and Supplementary Fig.
4). The pc-PET films hole-punched from these PET products were hydrolysed by serial treatment with FAST-PETase at 50 °C until the films were completely degraded (film disappeared). Enzyme
solution (200 nM of FAST-PETase in 100 mM KH2PO4-NaOH (pH 8.0) buffer) was replenished every 22 h. All measurements were conducted in triplicate (n = 3). The squares (mass loss) and circles
(PET monomers released) shown represent the individual numbers. The line connects mean values of the timepoints. Source data EXTENDED DATA FIG. 6 SCATTERPLOT OF TIME NEEDED FOR COMPLETE
DEGRADATION VERSUS INITIAL MASS OF THE HOLE-PUNCHED FILMS FROM 51 DIFFERENT PC-PET PRODUCTS. Degradation time was found to be corelated with the thickness (as thickness and mass are related)
of the hole-punched films from various plastic products. Source data EXTENDED DATA FIG. 7 SCATTERPLOT OF DEGRADATION RATE VERSUS (A.) INITIAL MASS, (B.) CRYSTALLINITY%, (C.) WEIGHT AVERAGE
MOLECULAR WEIGHT (MW), (D.) NUMBER AVERAGE MOLECULAR WEIGHT (MN), OR (E.) POLYDISPERSITY INDICES OF THE HOLE-PUNCHED FILMS FROM 51 DIFFERENT PC-PET PRODUCTS. Degradation rate was not found
to be dependent on any one metric across these various pc-PET plastics. Source data EXTENDED DATA FIG. 8 SCANNING ELECTRON MICROSCOPIC ANALYSIS OF THE PC-PET FILMS. The hole-punched PET
films from a bean cake PET container were treated with FAST-PETase for 0 h, 8 h, 16 h in 100 mM KH2PO4-NaOH (pH 8.0) buffer at 50 °C. EXTENDED DATA FIG. 9 DEPOLYMERIZATION OF THE
FINISH/NECK, BODY AND BASE CENTER FRAGMENTS OF AN UNTREATED WATER BOTTLE. Depolymerization was tested by FAST-PETase, wild-type PETase (WT), ThermoPETase (Thermo), DuraPETase (Dura), LCC and
ICCM at (A) 50 ºC, (B) 60 ºC, and (C) 72 ºC. All measurements were conducted in triplicate (n = 3). The bars and circles shown for each enzyme represent the average and individual numbers,
respectively. This comparative analysis provides two main conclusions. First, although higher reaction temperatures do promote the hydrolytic activity of the thermophilic LCC and ICCM
against the amorphous parts of the bottle (base center and finish), the highly crystalline body part still cannot be efficiently depolymerized by any tested enzymes and temperatures. Second,
FAST-PETase at 50 ºC exhibited the highest overall depolymerization rate seen in these experiments releasing 42, 0.14 and 15 mM of PET monomers within 24 h against the finish, body, and
bottom center of the bottle respectively. These values are 25%, 43% and 20% higher, respectively, than that of ICCM at 72 ºC. Source data SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION
This file contains Supplementary Methods, Discussion; Tables 1–3 and Figs. 1–13. REPORTING SUMMARY SOURCE DATA SOURCE DATA FIG. 1. SOURCE DATA FIG. 2. SOURCE DATA FIG. 3. SOURCE DATA FIG. 4.
SOURCE DATA EXTENDED DATA FIG. 5. SOURCE DATA EXTENDED DATA FIG. 6. SOURCE DATA EXTENDED DATA FIG. 7. SOURCE DATA EXTENDED DATA FIG. 9. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT
THIS ARTICLE CITE THIS ARTICLE Lu, H., Diaz, D.J., Czarnecki, N.J. _et al._ Machine learning-aided engineering of hydrolases for PET depolymerization. _Nature_ 604, 662–667 (2022).
https://doi.org/10.1038/s41586-022-04599-z Download citation * Received: 10 October 2021 * Accepted: 28 February 2022 * Published: 27 April 2022 * Issue Date: 28 April 2022 * DOI:
https://doi.org/10.1038/s41586-022-04599-z SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Danny Amendola Seemingly Takes a Shot at Zedd After Reports the DJ Might Be Dating Olivia CulpoDanny Amendola Seemingly Takes a Shot at Zedd After Reports the DJ Might Be Dating Olivia Culpo Culpo and Zedd recently ...
Epidural critic's anger is misplaced* So Sheryl Clark is “appalled” [“Epidurals’ Costs Should Not Be Paid,” Letters, June 27] because a class-action lawsuit...
Mr Cowie v Vesuvius plc and others: 2202735/2021 - GOV.UKMr Cowie v Vesuvius plc and others: 2202735/2021 Employment Tribunal decision. From: HM Courts & Tribunals Service and E...
Obama's 'drag queen' ornament | The WeekAndrew Breitbart's Big Government website posted photos of ornaments on a White House Christmas tree that feature images...
Prostate cancer: eat this to help prevent condition“Tomatoes may help prevent prostate cancer, as well as reduce tumour growth among men with the condition,” said medical ...
Latests News
Machine learning-aided engineering of hydrolases for pet depolymerizationABSTRACT Plastic waste poses an ecological challenge1,2,3 and enzymatic degradation offers one, potentially green and sc...
Defacing of van called hate crimeAn Antelope Valley church van was broken into and tagged with racist messages last week, vandalism that sheriff’s deputi...
Political will, people's determination can stop infiltration: rssOnly political will and determination of the people can help find a final solution to the vexed problem of infiltration ...
Surfing-Simmers, Chianca triumph in Portugal barrel-fest | WTVB | 1590 AM · 95.5 FM | The Voice of Branch CountyBy Lincoln Feast(Reuters) – Brazil’s Joao Chianca and American rookie Caitlin Simmers survived the slamming beachbreak b...
Band structure engineering of 2d materials using patterned dielectric superlatticesABSTRACT The ability to manipulate electrons in two-dimensional materials with external electric fields provides a route...