The structure of genotype-phenotype maps makes fitness landscapes navigable
The structure of genotype-phenotype maps makes fitness landscapes navigable"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
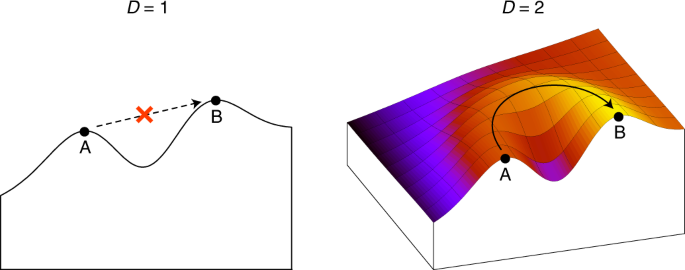
ABSTRACT Fitness landscapes are often described in terms of ‘peaks’ and ‘valleys’, indicating an intuitive low-dimensional landscape of the kind encountered in everyday experience. The space
of genotypes, however, is extremely high dimensional, which results in counter-intuitive structural properties of genotype-phenotype maps. Here we show that these properties, such as the
presence of pervasive neutral networks, make fitness landscapes navigable. For three biologically realistic genotype-phenotype map models—RNA secondary structure, protein tertiary structure
and protein complexes—we find that, even under random fitness assignment, fitness maxima can be reached from almost any other phenotype without passing through fitness valleys. This in turn
indicates that true fitness valleys are very rare. By considering evolutionary simulations between pairs of real examples of functional RNA sequences, we show that accessible paths are also
likely to be used under evolutionary dynamics. Our findings have broad implications for the prediction of natural evolutionary outcomes and for directed evolution. Access through your
institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio
journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 digital issues and online access to articles
$119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are
calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
EVOLVABILITY-ENHANCING MUTATIONS IN THE FITNESS LANDSCAPES OF AN RNA AND A PROTEIN Article Open access 19 June 2023 ROBUSTNESS AND INNOVATION IN SYNTHETIC GENOTYPE NETWORKS Article Open
access 28 April 2023 ENVIRONMENTAL SELECTION AND EPISTASIS IN AN EMPIRICAL PHENOTYPE–ENVIRONMENT–FITNESS LANDSCAPE Article 24 February 2022 DATA AVAILABILITY The dataset containing fRNA
(fRNAdb) used in this paper is available at: https://doi.org/10.18908/lsdba.nbdc00452-001. The GP maps analysed are available in the Code availability section. CODE AVAILABILITY The
ViennaRNA package (v.1.8.5), RNAshape package https://anaconda.org/bioconda/rnashapes and custom C++ and Python source code was used to construct GP maps and perform computational
simulations. The source code is available at: https://github.com/sgreenbury/gp-maps-nav. REFERENCES * Wright, S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution.
In _Proc. 6th International Congress on Genetics_ Vol. 1, 356–366 (1932). * Kauffman, S. A. _The Origins of Order: Self-Organization and Selection in Evolution_ (Oxford Univ. Press, 1993).
* Svensson, E. & Calsbeek, R. _The Adaptive Landscape in Evolutionary Biology_ (Oxford Univ. Press, 2012). * Pigliucci, M. in _The Adaptive Landscape in Evolutionary Biology_ (eds
Svensson, E. & Calsbeek, R.) 26–38 (Oxford Univ. Press, 2012). * Arjan, J., de Visser, G. M. & Krug, J. Empirical fitness landscapes and the predictability of evolution. _Nat. Rev.
Genetics_ 15, 480–490 (2014). Article Google Scholar * Fragata, Inês, Blanckaert, A., António Dias Louro, M., Liberles, D. A. & Bank, C. Evolution in the light of fitness landscape
theory. _Trends Ecol. Evol._ 34, 69–82 (2019). Article PubMed Google Scholar * Fisher, R. A. _The Genetical Theory of Natural Selection_ (Clarendon Press, 1958). * May, R. M. _Stability
and Complexity in Model Ecosystems_ (Princeton Univ. Press, 1973). * Conrad, M. & Ebeling, W. M.V. Volkenstein, evolutionary thinking and the structure of fitness landscapes.
_BioSystems_ 27, 125–128 (1992). Article CAS PubMed Google Scholar * Gavrilets, S. _Fitness Landscapes and the Origin of Species (MPB-41)_ (Princeton Univ. Press, 2004). * Franke, J.,
Klözer, A., Arjan, J., de Visser, G. M. & Krug, J. Evolutionary accessibility of mutational pathways. _PLoS Comput. Biol._ 7, e1002134 (2011). Article CAS PubMed PubMed Central
Google Scholar * Das, S. G., Direito, SusanaO. L., Waclaw, B., Allen, R. J. & Krug, J. Predictable properties of fitness landscapes induced by adaptational tradeoffs. _eLife_ 9, 1–24
(2020). Article Google Scholar * Weinreich, D. M., Watson, R. A. & Chao, L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. _Evolution_ 59, 1165–1174
(2005). CAS PubMed Google Scholar * Carneiro, Maurício & Hartl, D. L. Adaptive landscapes and protein evolution. _Proc. Natl Acad. Sci. USA_ 107, 1747–1751 (2010). Article CAS
PubMed PubMed Central Google Scholar * Wu, N. C., Dai, L., Olson, C. A., Lloyd-Smith, J. O. & Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths.
_eLife_ 5, 1–21 (2016). Article Google Scholar * Bank, C., Matuszewski, S., Hietpas, R. T. & Jensen, J. D. On the (un)predictability of a large intragenic fitness landscape. _Proc.
Natl Acad. Sci. USA_ 113, 14085–14090 (2016). Article CAS PubMed PubMed Central Google Scholar * Aguilar-Rodríguez, José, Payne, J. L. & Wagner, A. A thousand empirical adaptive
landscapes and their navigability. _Nat. Ecol. Evol._ 1, 0045 (2017). Article Google Scholar * Domingo, J. úlia, Diss, G. & Lehner, B. Pairwise and higher-order genetic interactions
during the evolution of a tRNA. _Nature_ 558, 117–121 (2018). Article CAS PubMed PubMed Central Google Scholar * Zheng, J., Payne, J. L. & Wagner, A. Cryptic genetic variation
accelerates evolution by opening access to diverse adaptive peaks. _Science_ 365, 347–353 (2019). Article CAS PubMed Google Scholar * Pokusaeva, V. O. et al. An experimental assay of the
interactions of amino acids from orthologous sequences shaping a complex fitness landscape. _PLOS Genet._ 15, e1008079 (2019). Article CAS PubMed PubMed Central Google Scholar *
Wagner, A. _Life Finds a Way: What Evolution Teaches Us about Creativity_ (Oneworld Publications, 2019). * Poelwijk, F. J., Kiviet, D. J., Weinreich, D. M. & Tans, S. J. Empirical
fitness landscapes reveal accessible evolutionary paths. _Nature_ 445, 383–386 (2007). Article CAS PubMed Google Scholar * Lobkovsky, A. E. & Koonin, E. V. Replaying the tape of
life: quantification of the predictability of evolution. _Front. Genet._ 3, 246 (2012). Article PubMed PubMed Central Google Scholar * Hartl, D. L. What can we learn from fitness
landscapes? _Curr. Opin. Microbiol._ 21, 51–57 (2014). Article PubMed Google Scholar * Louis, A. A. Contingency, convergence and hyper-astronomical numbers in biological evolution. _Stud.
Hist. Philos. Sci. C. Stud. Hist. Philos. Biol. Biomed. Sci._ 58, 107–116 (2016). Article Google Scholar * Kauffman, S. & Levin, S. Towards a general theory of adaptive walks on
rugged landscapes. _J. Theo. Biol._ 128, 11–45 (1987). Article CAS Google Scholar * Zagorski, M., Burda, Z. & Waclaw, B. Beyond the hypercube: evolutionary accessibility of fitness
landscapes with realistic mutational networks. _PLoS Comput. Biol._ 12, e1005218 (2016). Article PubMed PubMed Central Google Scholar * Kingman, J. F. C. A simple model for the balance
between selection and mutation. _J. Appl. Probab_. https://doi.org/10.2307/3213231 (1978). * Østman, B. & Adami, C. in _Recent Advances in the Theory and Application of Fitness
Landscapes_ (eds Richter, H. & Engelbrecht, A.) 509–526 (Springer, 2014). * Greenbury, S. F., Schaper, S., Ahnert, S. E. & Louis, A. A. Genetic correlations greatly increase
mutational robustness and can both reduce and enhance evolvability. _PLoS Comput. Biol._ 12, 1–27 (2016). Article Google Scholar * Ahnert, S. E. Structural properties of genotype–phenotype
maps. _J. R. Soc. Int._ 14, 20170275 (2017). Article Google Scholar * Manrubia, S. et al. From genotypes to organisms: state-of-the-art and perspectives of a cornerstone in evolutionary
dynamics. _Phys. Life Rev._ 38, 55–106 (2021). Article CAS PubMed Google Scholar * van Nimwegen, E., Crutchfield, J. P. & Huynen, M. Neutral evolution of mutational robustness.
_Proc. Natl Acad. Sci. USA_ 96, 9716–9720 (1999). Article PubMed PubMed Central Google Scholar * Greenbury, S. F. & Ahnert, S. E. The organization of biological sequences into
constrained and unconstrained parts determines fundamental properties of genotype-phenotype maps. _J. R. Soc. Interface_ 12, 20150724 (2015). Article CAS PubMed PubMed Central Google
Scholar * Manrubia, S. & Cuesta, JoséA. Distribution of genotype network sizes in sequence-to-structure genotype-phenotype maps. _J. R. Soc. Interface_ 14, 20160976 (2017). Article
PubMed PubMed Central Google Scholar * Weiß, M. & Ahnert, S. E. Phenotypes can be robust and evolvable if mutations have non-local effects on sequence constraints. _J. R. Soc.
Interface_ 15, 20170618 (2018). Article PubMed PubMed Central Google Scholar * Camargo, C. Q. & Louis, A. A. in _Complex Networks XI_ (eds Barbosa, H. et al.) 143–155 (Springer,
2020). * Wagner, A. Robustness and evolvability: a paradox resolved. _Proc. R. Soc. B: Bio. Sci._ 275, 91–100 (2008). Article Google Scholar * Schaper, S., Johnston, I. G. & Louis, A.
A. Epistasis can lead to fragmented neutral spaces and contingency in evolution. _Proc. R. Soc. B: Bio. Sci._ 279, 1777–1783 (2012). Article CAS Google Scholar * Kin, T. et al. fRNAdb: a
platform for mining/annotating functional RNA candidates from non-coding RNA sequences. _Nucleic Acids Res._ 35, 145–148 (2007). Article Google Scholar * Schuster, P., Fontana, W.,
Stadler, P. F. & Hofacker, I. L. From sequences to shapes and back: a case study in RNA secondary structures. _Proc. R. Soc. B. Bio. Sci._ 255, 279–284 (1994). Article CAS Google
Scholar * Hofacker, I. L. et al. Fast folding and comparison of RNA secondary structures. _Monatshefte Chem._ 125, 167–188 (1994). Article CAS Google Scholar * Fontana, W. Modelling
‘evo-devo’ with RNA. _BioEssays_ 24, 1164–1177 (2002). Article CAS PubMed Google Scholar * Cowperthwaite, M. C., Economo, E. P., Harcombe, W. R., Miller, E. L. & Meyers, LaurenAncel
The ascent of the abundant: how mutational networks constrain evolution. _PLoS Comput. Biol._ 4, e1000110 (2008). Article PubMed PubMed Central Google Scholar * Aguirre, J., Buldú, J.
M., Stich, M. & Manrubia, S. C. Topological structure of the space of phenotypes: the case of RNA neutral networks. _PLoS ONE_ 6, e26324 (2011). Article CAS PubMed PubMed Central
Google Scholar * Schaper, S. & Louis, A. A. The arrival of the frequent: how bias in genotype-phenotype maps can steer populations to local optima. _PLoS ONE_ 9, e86635 (2014). Article
PubMed PubMed Central Google Scholar * Wagner, A. _The Origins of Evolutionary Innovations: A Theory of Transformative Change in Living Systems_ (Oxford Univ. Press, 2011). * Greenbury,
S. F., Johnston, I. G., Louis, A. A. & Ahnert, S. E. A tractable genotype–phenotype map modelling the self-assembly of protein quaternary structure. _J. R. Soc. Interface_ 11, 20140249
(2014). Article PubMed PubMed Central Google Scholar * Johnston, I. G. et al. Symmetry and simplicity spontaneously emerge from the algorithmic nature of evolution. _Proc. Natl Acad.
Sci. USA_ 119, e2113883119 (2022). Article CAS PubMed PubMed Central Google Scholar * Dill, K. A. Theory for the folding and stability of globular proteins. _Biochemistry_ 24, 1501–1509
(1985). Article CAS PubMed Google Scholar * Irbäck, A. & Troein, C. Enumerating designing sequences in the HP model. _J. Biol. Phys._ 28, 1–15 (2002). Article PubMed PubMed
Central Google Scholar * Ferrada, E. & Wagner, A. A comparison of genotype-phenotype maps for RNA and proteins. _Biophysical J._ 102, 1916–1925 (2012). Article CAS Google Scholar *
Jörg, T., Martin, O. & Wagner, A. Neutral network sizes of biological RNA molecules can be computed and are not atypically small. _BMC Bioinformatics_ 9, 464 (2008). Article PubMed
PubMed Central Google Scholar * Borg, I. & Groenen, P. J. F. _Modern Multidimensional Scaling: Theory and Applications_ (Springer Science & Business Media, 2005). * Dingle, K.,
Ghaddar, F., Šulc, P. & Louis, A. A. Phenotype bias determines how natural RNA structures occupy the morphospace of all possible shapes. _Mol. Biol. Evol._ 39, msab280 (2021). * Dingle,
K., Schaper, S. & Louis, A. A. The structure of the genotype-phenotype map strongly constrains the evolution of non-coding RNA. _Int. Focus_ 5, 20150053 (2015). Google Scholar *
McCandlish, D. M. & Stoltzfus, A. Modeling evolution using the probability of fixation: history and implications. _Quart. Rev. Biol._ 89, 225–252 (2014). Article PubMed Google Scholar
* Kimura, M. On the probability of fixation of mutant genes in a population. _Genetics_ 47, 713–719 (1962). Article CAS PubMed PubMed Central Google Scholar * Giegerich, R., Voß,
Björn & Rehmsmeier, M. Abstract shapes of RNA. _Nucleic Acids Res._ 32, 4843–4851 (2004). Article CAS PubMed PubMed Central Google Scholar * Martin, N. S. & Ahnert, S. E.
Insertions and deletions in the RNA sequence-structure map. _J. R. Soc. Interface_ 18, 20210380 (2021). Article CAS PubMed PubMed Central Google Scholar * Catalán, P., Arias, C. F.,
Cuesta, J. A. & Manrubia, S. Adaptive multiscapes: an up-to-date metaphor to visualize molecular adaptation. _Biol. Direct_ 12, 7 (2017). Article PubMed PubMed Central Google Scholar
* Ogbunugafor, C. B., Wylie, C. S., Diakite, I., Weinreich, D. M. & Hartl, D. L. Adaptive landscape by environment interactions dictate evolutionary dynamics in models of drug
resistance. _PLoS Comput. Biol_. https://doi.org/10.1371/journal.pcbi.1004710 (2016). * Nichol, D., Robertson-Tessi, M., Anderson, AlexanderR. A. & Jeavons, P. Model genotype-phenotype
mappings and the algorithmic structure of evolution. _J. R. Soc. Interface_ 16, 20190332 (2019). Article CAS PubMed PubMed Central Google Scholar * Diaz-Uriarte, R. Cancer progression
models and fitness landscapes: a many-to-many relationship. _Bioinformatics_ 34, 836–844 (2018). Article CAS PubMed Google Scholar * Gabbutt, C. & Graham, T. A. Evolution’s
cartographer: mapping the fitness landscape in cancer. _Cancer Cell_ 39, 1311–1313 (2021). Article CAS PubMed Google Scholar * Lau, KitFun & Dill, K. A. A lattice statistical
mechanics model of the conformational and sequence spaces of proteins. _Macromolecules_ 22, 3986–3997 (1989). Article CAS Google Scholar * Li, H., Helling, R., Tang, C. & Wingreen, N.
Emergence of preferred structures in a simple model of protein folding. _Science_ 273, 666–669 (1996). Article CAS PubMed Google Scholar * Weinreich, D. M. Darwinian evolution can
follow only very few mutational paths to fitter proteins. _Science_ 312, 111–114 (2006). Article CAS PubMed Google Scholar * Ewens, W. J. _Mathematical Population Genetics: Theoretical
Introduction_ Vol. 1 (Springer, 2004). * Imhof, L. A. & Nowak, M. A. Evolutionary game dynamics in a Wright-Fisher process. _J. Math. Biol._ 52, 667–681 (2006). Article PubMed PubMed
Central Google Scholar Download references ACKNOWLEDGEMENTS S.E.A. was supported by the Royal Society and the Gatsby Foundation. S.F.G. was supported by the Engineering and Physical
Sciences Research Council. We thank M. Weiß for helpful discussions and insights. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Theory of Condensed Matter Group, Cavendish Laboratory,
University of Cambridge, Cambridge, UK Sam F. Greenbury * The Alan Turing Institute, British Library, London, UK Sam F. Greenbury & Sebastian E. Ahnert * Rudolf Peierls Centre for
Theoretical Physics, University of Oxford, Oxford, UK Ard A. Louis * Department of Chemical Engineering and Biotechnology, University of Cambridge, Cambridge, UK Sebastian E. Ahnert Authors
* Sam F. Greenbury View author publications You can also search for this author inPubMed Google Scholar * Ard A. Louis View author publications You can also search for this author inPubMed
Google Scholar * Sebastian E. Ahnert View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS S.F.G., A.A.L. and S.E.A. conceived and designed the
experiments. S.F.G. performed the experiments. S.F.G., A.A.L. and S.E.A. analysed the data. S.E.A. supervised the work. S.F.G., A.A.L. and S.E.A. wrote the paper. CORRESPONDING AUTHORS
Correspondence to Sam F. Greenbury, Ard A. Louis or Sebastian E. Ahnert. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW
INFORMATION _Nature Ecology & Evolution_ thanks Jacobo Aguirre and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. ADDITIONAL INFORMATION
PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 DEPICTION OF THE
DIFFERENT BIOLOGICAL SYSTEMS, SPECIFIC GP MAPS CONSIDERED AND EXAMPLE GENOTYPE, PHENOTYPE AND ENCODING OF PHENOTYPE. Each row is a specific GP map included in this work and is situated
within one of the four categories of system: RNA, Polyomino, HP (compact), and HP (non-compact). RNA and HP genotypes are depicted with distinct colours for their constituent bases.
Polyomino genotypes are shown as numerical sequences that map to the edges of distinctly coloured tiles with arrows used to indicate the tile orientation. The corresponding phenotype (the
structure that is formed following the self-assembly process on the example genotype) is shown with the colours and arrows used in the genotype depiction highlighting the mechanism by which
bonds are formed. The encoding of the example phenotypes are shown in the final column: dot-bracket and shape notation for RNA, grid coordinates for tile placements of polyominoes, and the
lattice directions for the HP lattice fold. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. REPORTING SUMMARY. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE
THIS ARTICLE Greenbury, S.F., Louis, A.A. & Ahnert, S.E. The structure of genotype-phenotype maps makes fitness landscapes navigable. _Nat Ecol Evol_ 6, 1742–1752 (2022).
https://doi.org/10.1038/s41559-022-01867-z Download citation * Received: 21 September 2021 * Accepted: 01 August 2022 * Published: 29 September 2022 * Issue Date: November 2022 * DOI:
https://doi.org/10.1038/s41559-022-01867-z SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Rugby legend bryan habana pens emotional statement as he announces his retirement - ruckSPRINGBOK LEGEND BRYAN HABANA HAS ANNOUNCED HIS RETIREMENT FROM ALL RUGBY AND WILL HANG UP HIS BOOTS AT THE END OF THE C...
Patricia GUZMAN | Premiere.frBiographie News Photos Vidéos Films Séries Nom de naissance GUZMAN Avis PoorNot so pooraveragegoodvery good Filmographie...
Should practitioners override patient choices? | nursing timesPatients sometimes make choices that may harm their health. Should practitioners be able to override these choices if th...
Page Not Found很抱歉,你所访问的页面已不存在了。如有疑问,请电邮[email protected]你仍然可选择浏览首页或以下栏目内容 :新闻生活娱乐财经体育视频播客新报业媒体有限公司版权所有(公司登记号:202120748H)...
Why justine bateman will never get plastic surgery: 'stop telling women to get their faces fixed'When Justine Bateman was in her early 40s and writing her first book _Fame: The Hijacking of Reality,_ she remembers Goo...
Latests News
The structure of genotype-phenotype maps makes fitness landscapes navigableABSTRACT Fitness landscapes are often described in terms of ‘peaks’ and ‘valleys’, indicating an intuitive low-dimension...
Jeremy guscott selects his england xv to face all blacks, genge and curry miss out - ruckI_N A PRE-SERIES SELECTION FOR THE RUGBY PAPER, ENGLAND LEGEND JEREMY GUSCOTT NAMED HIS IDEAL LINEUP TO FACE THE ALL BLA...
Ease seasonal depression the easy way with valkee ear buds | techcrunchI’ve always worried about people on depression meds; there are all kinds of meds, like Prozac and whatnot, then if that’...
Something went wrong, sorry. :(बॉलीवुड सुपरस्टार सलमान खान भले ही जेल में हों। लेकिन इससे उनके फैंस की संख्या में कोई कमी नहीं आई है। उल्टा वे सलमान के...
Something went wrong, sorry. :(Nace no seo dunha familia burguesa, iniciando os seus estudos nun colexio xesuíta.Comeza a carreira de Dereito, que aban...