Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance
Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
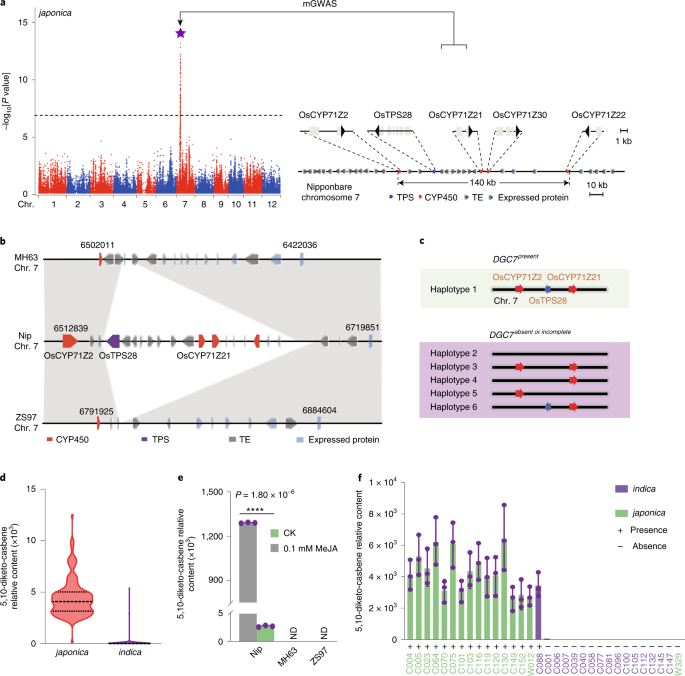
ABSTRACT Diterpenoids are the major group of antimicrobial phytoalexins in rice1,2. Here, we report the discovery of a rice diterpenoid gene cluster on chromosome 7 (_DGC7_) encoding the
entire biosynthetic pathway to 5,10-diketo-casbene, a member of the monocyclic casbene-derived diterpenoids. We revealed that _DGC7_ is regulated directly by JMJ705 through methyl
jasmonate-mediated epigenetic control3. Functional characterization of pathway genes revealed _OsCYP71Z21_ to encode a casbene C10 oxidase, sought after for the biosynthesis of an array of
medicinally important diterpenoids. We further show that _DGC7_ arose relatively recently in the _Oryza_ genus, and that it was partly formed in _Oryza_ _rufipogon_ and positively selected
for in _japonica_ during domestication. Casbene-synthesizing enzymes that are functionally equivalent to OsTPS28 are present in several species of Euphorbiaceae but gene tree analysis shows
that these and other casbene-modifying enzymes have evolved independently. As such, combining casbene-modifying enzymes from these different families of plants may prove effective in
producing a diverse array of bioactive diterpenoid natural products. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution
ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time
Learn more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS CHROMOSOME-SCALE PEARL MILLET GENOMES REVEAL _CLAMT1B_ AS KEY DETERMINANT OF STRIGOLACTONE
PATTERN AND STRIGA SUSCEPTIBILITY Article Open access 12 August 2024 GENOME-WIDE CHARACTERIZATION OF THE SORGHUM _JAZ_ GENE FAMILY AND THEIR RESPONSES TO PHYTOHORMONE TREATMENTS AND APHID
INFESTATION Article Open access 25 February 2022 GENOME-WIDE INVESTIGATION OF CYTOKININ OXIDASE/DEHYDROGENASE (CKX) FAMILY GENES IN _BRASSICA JUNCEA_ WITH AN EMPHASIS ON YIELD-INFLUENCING
_CKX_ Article Open access 29 May 2025 DATA AVAILABILITY The sequences data of 424 O_. sativa_ accessions are available in the National Center for Biotechnology Information (NCBI) under the
BioProject PRJNA171289 (ref. 17). The single nucleotide polymorphisms information of 424 _O. sativa_ accessions is available in RiceVarMap (http://ricevarmap.ncpgr.cn/v1). The pangenome data
were acquired from the pangenome dataset (https://figshare.com/collections/Novel_sequences_structural_variations_and_gene_presence_variations_of_Asian_cultivated_rice/3876022/1 and
http://cgm.sjtu.edu.cn/3kricedb/)18,19,24. Thirteen accessions of _O. rufipogon_ were selected from 446 diverse _O. rufipogon_ accessions from Asia and Oceania, and represented all the major
genetically distinct clusters in _O. rufipogon_; the other ten wild rice are from EnsemblPlants (http://plants.ensembl.org/index.html) and National Genomics Data Center
(https://bigd.big.ac.cn/search?dbId=gwh&q=Oryza), including _O. barthii_ (AA), _O. glumipatula_ (AA), _O. glaberrima_ (AA), _O. meridionalis_ (AA), _O. longistaminata_ (AA), _O. nivara_
(AA), _O. brachyantha_ (FF), _O. punctata_ (BB) and _O. brachyantha_ (GG)24. Genes reported in the study are deposited in the NCBI. The genes can be found in GenBank or Rice Genome
Annotation Project database (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) under the following accession numbers: OsTPS28, MN833254; OsCYP71Z21, LOC_Os07g11870; OsCYP71Z2,
LOC_Os07g11739; OsCYP71Z22, LOC_Os07g11970; OsCYP71Z30, LOC_Os07g11890. Source data are provided with this paper. CHANGE HISTORY * _ 16 DECEMBER 2020 A Correction to this paper has been
published: https://doi.org/10.1038/s41477-020-00838-1. _ REFERENCES * Schmelz, E. A. et al. Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. _Plant J._ 79, 659–678
(2014). CAS PubMed Google Scholar * Lu, X. et al. Inferring roles in defense from metabolic allocation of rice diterpenoids. _Plant Cell_ 30, 1119–1131 (2018). CAS PubMed PubMed Central
Google Scholar * Li, T. et al. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. _Plant Cell_
25, 4725–4736 (2013). CAS PubMed PubMed Central Google Scholar * Chen, X. et al. Biological activities and potential molecular targets of cucurbitacins: a focus on cancer. _Anti-Cancer
Drug_ 23, 777–787 (2012). CAS Google Scholar * Huang, A. C. et al. A specialized metabolic network selectively modulates _Arabidopsis_ root microbiota. _Science_ 364, eaau6389 (2019). CAS
PubMed Google Scholar * Nützmann, H.-W., Huang, A. & Osbourn, A. Plant metabolic clusters—from genetics to genomics. _New Phytol._ 211, 771–789 (2016). PubMed PubMed Central Google
Scholar * Zi, J., Mafu, S. & Peters, R. J. To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. _Annu. Rev. Plant Biol._ 65, 259–286 (2014). CAS PubMed
PubMed Central Google Scholar * Panizza, B. J. et al. Phase I dose-escalation study to determine the safety, tolerability, preliminary efficacy and pharmacokinetics of an intratumoral
injection of tigilanol tiglate (EBC-46). _EBioMedicine_ 50, 433–441 (2019). PubMed PubMed Central Google Scholar * Hezareh, M. Prostratin as a new therapeutic agent targeting HIV viral
reservoirs. _Drug News Perspect._ 18, 496–500 (2005). CAS PubMed Google Scholar * Johnson, H. E., Banack, S. A. & Cox, P. A. Variability in content of the anti-AIDS drug candidate
prostratin in Samoan populations of _Homalanthus nutans_. _J. Nat. Prod._ 71, 2041–2044 (2008). CAS PubMed PubMed Central Google Scholar * Lebwohl, M. et al. Ingenol mebutate gel for
actinic keratosis. _N. Engl. J. Med._ 366, 1010–1019 (2012). CAS PubMed Google Scholar * Sabandar, C. W., Ahmat, N., Jaafar, F. M. & Sahidin, I. Medicinal property, phytochemistry and
pharmacology of several _Jatropha_ species (Euphorbiaceae): a review. _Phytochemistry_ 85, 7–29 (2013). CAS PubMed Google Scholar * Inoue, Y. et al. Identification of a novel
casbane-type diterpene phytoalexin, _ent_-10-oxodepressin, from rice leaves. _Biosci. Biotech. Biochem._ 77, 760–765 (2013). CAS Google Scholar * Horie, K., Sakai, K., Okugi, M., Toshima,
H. & Hasegawa, M. Ultraviolet-induced amides and casbene diterpenoids from rice leaves. _Phytochem. Lett._ 15, 57–62 (2016). CAS Google Scholar * Li, W. et al. OsCYP71Z2 involves
diterpenoid phytoalexin biosynthesis that contributes to bacterial blight resistance in rice. _Plant Sci._ 207, 98–107 (2013). CAS PubMed Google Scholar * Li, W. et al. Overexpressing
CYP71Z2 enhances resistance to bacterial blight by suppressing auxin biosynthesis in rice. _PLoS ONE_ 10, e0119867 (2015). PubMed PubMed Central Google Scholar * Zhao, H. et al.
RiceVarMap: a comprehensive database of rice genomic variations. _Nucleic Acids Res._ 43, D1018–D1022 (2015). CAS PubMed Google Scholar * Wang, W. et al. Genomic variation in 3,010
diverse accessions of Asian cultivated rice. _Nature_ 557, 43–49 (2018). CAS PubMed PubMed Central Google Scholar * Huang, X. et al. A map of rice genome variation reveals the origin of
cultivated rice. _Nature_ 490, 497–501 (2012). CAS PubMed PubMed Central Google Scholar * Chen, W. et al. Genome-wide association analyses provide genetic and biochemical insights into
natural variation in rice metabolism. _Nat. Genet._ 46, 714–721 (2014). CAS PubMed Google Scholar * Yu, N. et al. Delineation of metabolic gene clusters in plant genomes by chromatin
signatures. _Nucleic Acids Res._ 44, 2255–2265 (2016). CAS PubMed PubMed Central Google Scholar * Zhou, S. et al. Cooperation between the H3K27me3 chromatin mark and non-CG methylation
in epigenetic regulation. _Plant Physiol._ 172, 1131–1141 (2016). CAS PubMed PubMed Central Google Scholar * Nützmann, H.-W. et al. Active and repressed biosynthetic gene clusters have
spatially distinct chromosome states. _Proc. Natl Acad. Sci. USA_ 24, 13800–13809 (2020). Google Scholar * Stein, J. C. et al. Genomes of 13 domesticated and wild rice relatives highlight
genetic conservation, turnover and innovation across the genus _Oryza_. _Nat. Genet._ 50, 285–296 (2018). CAS PubMed Google Scholar * Wing, R. A., Purugganan, M. D. & Zhang, Q. The
rice genome revolution: from an ancient grain to green super rice. _Nat. Rev. Genet._ 19, 505–517 (2018). CAS PubMed Google Scholar * King, A. J., Brown, G. D., Gilday, A. D., Larson, T.
R. & Graham, I. A. Production of bioactive diterpenoids in the Euphorbiaceae depends on evolutionarily conserved gene clusters. _Plant Cell_ 26, 3286–3298 (2014). CAS PubMed PubMed
Central Google Scholar * Takos, A. M. et al. Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in _Lotus japonicus_ and suggests the repeated
evolution of this chemical defence pathway. _Plant J._ 68, 273–286 (2011). CAS PubMed Google Scholar * Field, B. & Osbourn, A. E. Metabolic diversification—independent assembly of
operon-like gene clusters in different plants. _Science_ 320, 543–547 (2008). CAS PubMed Google Scholar * Wang, M. et al. Parallel selection on a dormancy gene during domestication of
crops from multiple families. _Nat. Genet._ 50, 1435–1441 (2018). CAS PubMed Google Scholar * Swaminathan, S., Morrone, D., Wang, Q., Fulton, D. B. & Peters, R. J. CYP76M7 is an
_ent_-cassadiene C11-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. _Plant Cell_ 21, 3315–3325 (2009). CAS PubMed PubMed Central Google
Scholar * Wang, Q., Hillwig, M. L. & Peters, R. J. CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. _Plant J._ 65, 87–95
(2011). CAS PubMed Google Scholar * Wang, Q. et al. Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. _J. Biol. Chem._ 287, 6159–6168
(2012). CAS PubMed PubMed Central Google Scholar * Miyamoto, K. et al. Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. _Plant J._ 87, 293–304 (2016). CAS
PubMed Google Scholar * Gross, B. L. & Zhao, Z. Archaeological and genetic insights into the origins of domesticated rice. _Proc. Natl Acad. Sci. USA_ 111, 6190–6197 (2014). CAS
PubMed PubMed Central Google Scholar * Qi, Z. Genetics and improvement of bacterial blight resistance of hybrid rice in China. _Rice Sci._ 23, 111–119 (2009). Google Scholar * Kauffman,
H. E., Reddy, A. P. K., Hsieh, S. P. Y. & Merca, S. D. An improved technique for evaluating resistance of rice varieties to _Xanthomonas oryzae_. _Plant Dis. Rep._ 57, 537–541 (1973).
Google Scholar * Zhang, H. et al. Transposon-derived small RNA is responsible for modified function of WRKY45 locus. _Nat. Plants_ 2, 16016 (2016). CAS PubMed Google Scholar * Zhang, B.
et al. Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to _Xanthomonas oryzae_ pv. _oryzae_. _Plant Commun._ 1, 100088 (2020). PubMed
PubMed Central Google Scholar * Tao, Z. et al. A pair of allelic WRKY genes play opposite roles in rice–bacteria interactions. _Plant Physiol._ 151, 936–948 (2009). CAS PubMed PubMed
Central Google Scholar * King, A. J. et al. A cytochrome P450-mediated intramolecular carbon–carbon ring closure in the biosynthesis of multidrug-resistance-reversing lathyrane
diterpenoids. _ChemBioChem_ 17, 1593–1597 (2016). CAS PubMed PubMed Central Google Scholar * Boutanaev, A. M. et al. Investigation of terpene diversification across multiple sequenced
plant genomes. _Proc. Natl Acad. Sci. USA_ 112, E81–E88 (2015). CAS PubMed Google Scholar * Luo, D. et al. Oxidation and cyclization of casbene in the biosynthesis of Euphorbia factors
from mature seeds of _Euphorbia lathyris_ L. _Proc. Natl Acad. Sci. USA_ 113, E5082–E5089 (2016). CAS PubMed PubMed Central Google Scholar * Peng, M. et al. Differentially evolved
glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. _Nat. Commun._ 8, 1975 (2017). PubMed PubMed Central Google Scholar * Ikezawa, N. et al.
Lettuce costunolide synthase (CYP71BL2) and its homolog (CYP71BL1) from sunflower catalyze distinct regio- and stereoselective hydroxylations in sesquiterpene lactone metabolism. _J. Biol.
Chem._ 286, 21601–21611 (2011). CAS PubMed PubMed Central Google Scholar * Sainsbury, F., Thuenemann, E. C. & Lomonossoff, G. P. pEAQ: versatile expression vectors for easy and quick
transient expression of heterologous proteins in plants. _Plant Biotechnol. J._ 7, 682–693 (2009). CAS PubMed Google Scholar * Zeng, X. et al. Genome-wide dissection of co-selected UV-B
responsive pathways in the UV-B adaptation of qingke. _Mol. Plant_ 13, 112–127 (2020). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank J. D. Keasling, G. P.
Lomonossoff and Z. Zhao for their advice and their gift of the expression vectors and strains. We also thank D. R. Nelson, University of Tennessee, for help in naming the OsCYP71Z30. This
work was supported by the National Science Fund for Distinguished Young Scholars (grant no. 31625021), the State Key Programme of National Natural Science Foundation of China (grant no.
31530052) and the Hainan University Startup Fund KYQD(ZR)1866 to J.L. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * National Key Laboratory of Crop Genetic Improvement and National Center of
Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China Chuansong Zhan, Long Lei, Zixin Liu, Shen Zhou, Chenkun Yang, Xitong Zhu, Hao Guo, Feng Zhang, Meng Peng, Meng
Zhang, Yufei Li, Zixin Yang, Yangyang Sun, Yuheng Shi, Kang Li, Shuangqian Shen, Xuyang Wang, Jiawen Shao, Xinyu Jing, Zixuan Wang, Lianghuan Qu, Xianqing Liu, Ling-Ling Chen & Meng Yuan
* College of Tropical Crops, Hainan University, Haikou, China Long Lei, Hao Guo, Ling Liu, Xianqing Liu & Jie Luo * Centre for Novel Agricultural Products, Department of Biology,
University of York, York, UK Yi Li, Tomasz Czechowski & Ian Graham * College of Agriculture, Ibaraki University, Ami, Japan Morifumi Hasegawa * Graduate School of Biological Sciences,
Nara Institute of Science and Technology, Ikoma, Japan Takayuki Tohge * Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany Alisdair R. Fernie Authors * Chuansong Zhan
View author publications You can also search for this author inPubMed Google Scholar * Long Lei View author publications You can also search for this author inPubMed Google Scholar * Zixin
Liu View author publications You can also search for this author inPubMed Google Scholar * Shen Zhou View author publications You can also search for this author inPubMed Google Scholar *
Chenkun Yang View author publications You can also search for this author inPubMed Google Scholar * Xitong Zhu View author publications You can also search for this author inPubMed Google
Scholar * Hao Guo View author publications You can also search for this author inPubMed Google Scholar * Feng Zhang View author publications You can also search for this author inPubMed
Google Scholar * Meng Peng View author publications You can also search for this author inPubMed Google Scholar * Meng Zhang View author publications You can also search for this author
inPubMed Google Scholar * Yufei Li View author publications You can also search for this author inPubMed Google Scholar * Zixin Yang View author publications You can also search for this
author inPubMed Google Scholar * Yangyang Sun View author publications You can also search for this author inPubMed Google Scholar * Yuheng Shi View author publications You can also search
for this author inPubMed Google Scholar * Kang Li View author publications You can also search for this author inPubMed Google Scholar * Ling Liu View author publications You can also search
for this author inPubMed Google Scholar * Shuangqian Shen View author publications You can also search for this author inPubMed Google Scholar * Xuyang Wang View author publications You can
also search for this author inPubMed Google Scholar * Jiawen Shao View author publications You can also search for this author inPubMed Google Scholar * Xinyu Jing View author publications
You can also search for this author inPubMed Google Scholar * Zixuan Wang View author publications You can also search for this author inPubMed Google Scholar * Yi Li View author
publications You can also search for this author inPubMed Google Scholar * Tomasz Czechowski View author publications You can also search for this author inPubMed Google Scholar * Morifumi
Hasegawa View author publications You can also search for this author inPubMed Google Scholar * Ian Graham View author publications You can also search for this author inPubMed Google
Scholar * Takayuki Tohge View author publications You can also search for this author inPubMed Google Scholar * Lianghuan Qu View author publications You can also search for this author
inPubMed Google Scholar * Xianqing Liu View author publications You can also search for this author inPubMed Google Scholar * Alisdair R. Fernie View author publications You can also search
for this author inPubMed Google Scholar * Ling-Ling Chen View author publications You can also search for this author inPubMed Google Scholar * Meng Yuan View author publications You can
also search for this author inPubMed Google Scholar * Jie Luo View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.L. designed the research.
J.L., L.-L.C., L.Q., M.Y. and X.L. supervised this study. C.Z., L. Lei, S.Z., Z.L., F.Z., M.Z., Y. Sun, Y. Shi, K.L., T.C., M.H., I.G., Z.Y. and T.T. participated in the material
preparation. C.Z., C.Y., Yi Li, X.W. and J.S. carried out the metabolite analyses. C.Z., Z.L., S.Z., C.Y., X.Z., H.G., M.P., M.Z., Yufei Li, Z.Y., L. Liu, S.S., J.S., X.J., Yi Li, T.T. and
Z.W. performed the data analyses. C.Z., L. Lei, Z.L., S.Z. and C.Y. performed most of the experiments. J.L., C.Z., I.G. and A.R.F. wrote the manuscript. CORRESPONDING AUTHOR Correspondence
to Jie Luo. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Plants_ thanks the anonymous reviewers
for their contribution to the peer review of this work. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 THE DISTRIBUTION OF THE WORLDWIDE COLLECTION OF RICE ACCESSIONS IN THIS STUDY. The core collection of 424 cultivated rice accessions in this
study has a wide geographic distribution. Colour dots indicate different subspecies/type of cultivated rice. EXTENDED DATA FIG. 2 FUNCTIONAL ANALYSES OF OSTPS28, OSCYP71Z2 AND OSCYP71Z21. A,
Gas chromatography of the reaction products of OsTPS28 with GGDP. GGDP, geranylgeranyl diphosphate. Casbene and neocembrene reference compounds were purified from infiltrated _N.
benthamiana_ leaves by the method described previously26. Compound 1, casbene; Compound 2, neocembrene. B, Gas chromatography of _in vitro_ enzyme assays showing the 10-keto-casbene C5
oxidase activity of yeast-expressed CYP71Z2 in the present of NADPH. Microsomes prepared from yeast containing PESC-URA empty vector were used as a negative control. 10-keto-casbene
reference compound was purified from rice leaves by the method described previously13,14. Compound 3, 10-keto- casbene; Compound 4, 5,10-diketo-casbene. C, Gas chromatography of the extracts
prepared from the leaves of _N. benthamiana_ infiltrated with OsTPS28 overexpressing vector. EXTENDED DATA FIG. 3 MASS SPECTRUM AND STRUCTURE OF 5,10- DIKETO-CASBENE. A, Mass spectrum and
structure of the product in _N. benthamiana_ leaves simultaneously overexpressing _OsTPS28_, _OsCYP71Z2_ and _OsCYP71Z21_. B, Mass spectrum of 5,10-diketo-casbene reference. LC-MS, liquid
chromatography–mass spectrometry. C, 1H NMR (left) and 13C NMR (right) results of 5,10-diketo-casbene. EXTENDED DATA FIG. 4 THE EXPRESSION PROFILES OF GENES FROM _DGC7_. The genes from
_DGC7_ are indicated in bold. The transcript abundances of indicated genes in different organs at different stages were shown: expression levels of _OsTPS28_, _OsCYP17Z2_ and _OsCYP71Z21_ is
correlated at different developmental stages. The numerical values for blue-to-red gradient represent normalized expression levels from quantitative real-time PCR analysis. EXTENDED DATA
FIG. 5 THE CASBENE-TYPE DITERPENE BIOSYNTHESIS VIA DISTINCT BIOSYNTHETIC ROUTES IN RICE AND CASTOR. The casbene-type diterpene biosynthetic pathways in rice and castor. Chr.7, chromosome 7;
GGDP, geranylgeranyl diphosphate. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Methods, Figs. 1–16 and references. REPORTING SUMMARY SUPPLEMENTARY TABLES Supplementary
Tables 1–10. SOURCE DATA SOURCE DATA FIG. 1 Statistical source data. SOURCE DATA FIG. 3 Statistical source data. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Zhan, C., Lei, L., Liu, Z. _et al._ Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistance. _Nat. Plants_ 6, 1447–1454 (2020).
https://doi.org/10.1038/s41477-020-00816-7 Download citation * Received: 09 June 2020 * Accepted: 04 November 2020 * Published: 07 December 2020 * Issue Date: December 2020 * DOI:
https://doi.org/10.1038/s41477-020-00816-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Chance the Rapper Takes Some Heat For His Latest FestivalTough QuestionsChance the Rapper Takes Some Heat For His Latest FestivalThe star from Chicago responds to criticism of h...
AC/DC Reveal Retired Bassist Cliff Williams Will Return for Power Trip FestComebackAC/DC Reveal Retired Bassist Cliff Williams Will Return for Power Trip FestAhead of first concert since Septembe...
Lisa Tozzi – Rolling StoneLisa TozziExecutive Digital DirectorXemailLisa Tozzi is Rolling Stone’s Executive Digital Director. She oversees the dai...
NHS hospitals still using over 8,000 ‘archaic’ fax machinesNHS hospital trusts across England still own over 8,000 fax machines, despite the communications technology having large...
Eagles Turn Down Super Bowl, Prefer Grand Ole OpryMuch like the Philadelphia football team of the same name, Don Henley’s Eagles will not be playing at the Super Bowl nex...
Latests News
Selection of a subspecies-specific diterpene gene cluster implicated in rice disease resistanceABSTRACT Diterpenoids are the major group of antimicrobial phytoalexins in rice1,2. Here, we report the discovery of a r...
Attention Required! | CloudflarePlease enable cookies. Sorry, you have been blocked You are unable to access goodreturns.in Why have I been blocked? Thi...
Charles and wills surprise people queuing for queen’s coffinKate PurnellThe West Australian King Charles III and Prince William have made a surprise visit to the huge lines of peop...
Mr p weatherilt v cathay pacific airways ltd: ukeat/0333/16/rnMR P WEATHERILT V CATHAY PACIFIC AIRWAYS LTD: UKEAT/0333/16/RN Employment Appeal Tribunal judgment of Judge Richardson o...
Alan pardew's banking on massadio haidara returnThe left-back, 20, was on the wrong end of a reckless challenge by Wigan’s Callum McManaman two weeks ago. But fears tha...