Insihgt: an accessible multi-scale, multi-modal 3d spatial biology platform
Insihgt: an accessible multi-scale, multi-modal 3d spatial biology platform"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Biological systems are complex, encompassing intertwined spatial, molecular and functional features. However, methodological constraints limit the completeness of information that
can be extracted. Here, we report the development of INSIHGT, a non-destructive, accessible three-dimensional (3D) spatial biology method utilizing superchaotropes and host-guest chemistry
to achieve homogeneous, deep penetration of macromolecular probes up to centimeter scales, providing reliable semi-quantitative signals throughout the tissue volume. Diverse antigens, mRNAs,
neurotransmitters, and post-translational modifications are well-preserved and simultaneously visualized. INSIHGT also allows multi-round, highly multiplexed 3D molecular probing and is
compatible with downstream traditional histology and nucleic acid sequencing. With INSIHGT, we map undescribed podocyte-to-parietal epithelial cell microfilaments in mouse glomeruli and
neurofilament-intensive inclusion bodies in the human cerebellum, and identify NPY-proximal cell types defined by spatial morpho-proteomics in mouse hypothalamus. We anticipate that INSIHGT
can form the foundations for 3D spatial multi-omics technology development and holistic systems biology studies. SIMILAR CONTENT BEING VIEWED BY OTHERS THE EMERGING LANDSCAPE OF SPATIAL
PROFILING TECHNOLOGIES Article 20 July 2022 SM-OMICS IS AN AUTOMATED PLATFORM FOR HIGH-THROUGHPUT SPATIAL MULTI-OMICS Article Open access 10 February 2022 SPATIAL MAPPING OF PROTEIN
COMPOSITION AND TISSUE ORGANIZATION: A PRIMER FOR MULTIPLEXED ANTIBODY-BASED IMAGING Article 22 November 2021 INTRODUCTION The complexity of biological systems mandates high-dimensional
measurements to obtain an integrative understanding. However, measurements are inevitably perturbative, affecting the authenticity of the retrieved information. Spatially resolved
transcriptomics1 and highly multiplexed immunohistochemistry (IHC)2 have proven to be powerful approaches to extract spatial molecular insights from tissue slices, but the two-dimensional
(2D) readout limits the representativeness of the information extracted. Meanwhile, three-dimensional (3D) multiplexed visualization of tissue structural and molecular features can reveal
previously unknown organization principles3,4. Optical tissue clearing technologies promises to reveal the authentic 3D nature of tissue architecture and molecular distributions5. Despite
its significant advancements, the achievable depths of probe penetration limits the depth of analysis6. The limited penetration of antibodies in 3D IHC represents one of the most significant
barrier to 3D spatial biology6. In recent years, multiple creative solutions have been proposed for deep immunohistochemistry7,8,9,10. However, an accessible technology that balances the
authenticity and volume of data extracted is still lacking. For example, signal homogeneity across penetration depth is suboptimal with most methods, where probes preferentially deposit near
the tissue surface and complicates downstream quantitative protein expression determination6,11. The homogeneous penetration can only be attained either through complicated operations or
equipment11,12,13, or extensive tissue permeabilization14,15 or incubation times measuring in weeks8. These shortcomings hinder the wide adoption of 3D tissue analysis in research and
renders them unsatisfactory for clinical translation. Here, we report the development of In situ _H_ost-_G_uest Chemistry for _T_hree-dimensional Histology (INSIHGT). INSIHGT is a
user-friendly 3D histochemistry method, featuring (1) homogeneous probe penetration up to centimeter depths, (2) producing quantitative, highly specific immunostaining signals, (3) a fast
and affordable workflow to accommodate different tissue sizes and shapes, (4) simple immersion-based staining at room temperature, thus easily adopted in any laboratory and ready for scaling
and automate, and (5) uses off-the-shelf antibodies or probes and is directly applicable to otherwise unlabeled mouse and human tissues fixed with paraformaldehyde only. INSIHGT was
developed based on the manipulation of macromoleular diffusiophoresis using _closo_-dodecahydrododecaborate [B12H12]2-16 and a γ-cyclodextrin derivative. If tissue clearing is required,
INSIHGT works best with solvent-based clearing methods17,18,19. RESULTS MODULATION OF ANTIBODY-ANTIGEN BINDING FOR ENHANCED PROBE PENETRATION The limited penetration of macromolecular probes
in complex biological systems belongs to the broader subject of transport phenomena, where diffusion and advections respectively drive the dissipation and directional drift of mass, energy
and momentum. When biomolecules such as proteins are involved, the (bio)molecular fluxes are additionally determined by binding reactions, which can significantly deplete biomolecules due to
their high binding affinities and low concentrations employed - a “reaction barrier” to deep antibody penetration. This is first described and postulated by Renier et al.17 (as in
immunolabeling-enabled three-dimensional imaging of solvent-cleared organs, iDISCO) and Murray et al.14 (as in system-wide control of interaction time and kinetics of chemicals, SWITCH), and
the latter further showed that the modulation of antibody-antigen (Ab-Ag) binding affinity (SWITCH labeling) can lead to homogeneous penetration of up to 1 mm for an anti-Histone H3
antibody using low concentrations of sodium dodecyl sulfate (SDS). Other techniques similarly utilizes urea8, sodium deoxycholate12, and heat9 to modulate antibody-antigen binding. However,
others and we observed a general compromise between antibody labelling quality, penetration depth and uniformity, and duration of incubation. Deep penetration invariably requires long
incubation times with inhomogeneous signal across depth, while faster methods leads to weak or nonspecific staining, as well as non-uniform penetration8,9,17. Specifically, the use of SDS
for deep labelling with SWITCH labelling has only been demonstrated for a handful of antigens (e.g., Histone H314, NeuN20, ColIV, αSMA, and TubIII21). It was found that deep staining with
SDS was not universally applicable20, resulting in weak calbindin staining22, insufficient staining depth for β-amyloid plaques23, and often required tailored refinement of buffer
concentration24. In our validation data, we similarly observed the variable performance when SDS is co-applied with antibodies (Supplementary Fig. 1). Furthermore, although adding antibodies
or probes theoretically improves penetration via steep concentration gradients, either the cost becomes prohibitive or it produces a biased representation of rimmed surface staining
pattern6,8, especially for densely expressed binding targets. In the most extreme cases, the superficial staining signal would saturate microscope detectors while the core remains unstained
(Supplementary Fig. 2). Nonetheless, the conception of modulating antibody-antigen binding kinetics as a means to control probe flux through tissues is highly attractive12,14, given the
simplicity, scalability, and affordability should the method be robust and generalizable. We postulated that the reason for the highly variable performance of SDS-assisted deep
immunostaining is two-fold: the denaturation of antibodies beyond reparability, and the ineffective reinstatement of binding reactions. This prompted us to search for alternative approaches
that can tune biomolecular binding affinities while preserving both macromolecular probe mobility and stability. In addition, the negation of the modulatory effect should be efficient and
robust to reinstate biomolecular reactions within the complex tissue environment. Therefore, here we aim to develop a fast, equipment-free, deep and uniform multiplexed immunostaining
method, which will help bring 3D histology to any basic laboratories. BORON CLUSTER HOST–GUEST CHEMISTRY FOR IN SITU MACROMOLECULAR PROBE MOBILITY CONTROL Our initial attempts by using heat
and the GroEL-GroES system to denature and refold antibodies in situ respectively have proved unsuccessful (Supplementary Fig. 1). We thus switched from the natural molecular chaperones to
artificial ones using milder detergents (e.g., sodium deoxycholate (SDC) and 3-([3-Cholamidopropyl]dimethylammonio)- 2-hydroxy-1-propanesulfonate i.e., CHAPSO) and their
charge-complementary, size-matched host-complexing agents (e.g., β-cyclodextrins and their derivatives such as heptakis-(6-amino-6-deoxy)-beta-cyclodextrin, i.e., 6NβCD), which improved
antibody penetration and staining success rate (Supplementary Fig. 3). However, despite extensive optimization on the structure and derivatization on the detergents and their size- and
charge-complementary cyclodextrins, they still have limited generality for a panel of antibodies tested (Supplementary Fig. 3), producing nonspecific vascular precipitates or nuclear
stainings. We then explored the use of chaotropes, which are known to solubilize proteins with enhanced antibody penetration8. However, these approaches require long incubation times with
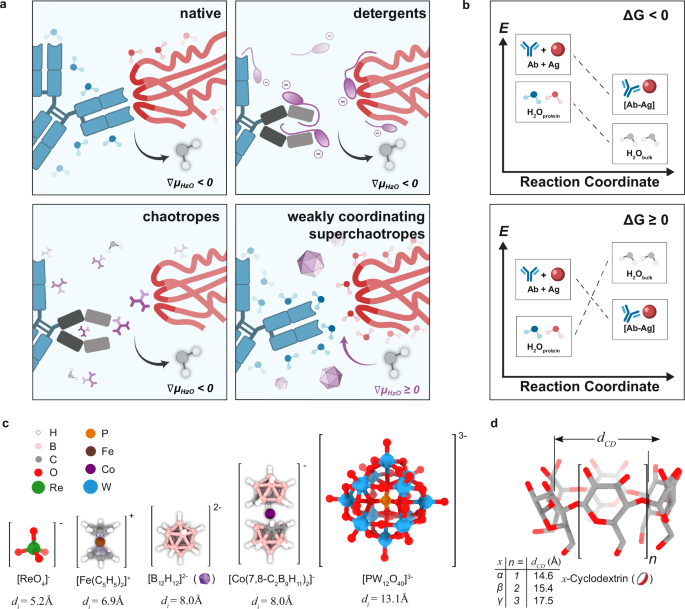
extensive tissue pre-processing. Furthermore, higher concentrations of chaotropes often denature proteins as they directly interact with various protein residues and backbone25,26 (Fig. 1a,
b). We hence focus on testing weakly coordinating superchaotropes (WCS), a class of chemicals that we hypothesized to inhibit antibody-antigen interactions while preserving their structure
and hence functions (Fig. 1a, b). We searched for weakly coordinating ions based on their utility in isolating extremely electrophilic species for X-ray crystallography, or as conjugate
bases of superacids. We can then select a subset of these coordinatively inert ionic species that possess high chaotropicity as candidates for our deep immunostaining purpose. After
antibodies and WCS have been homogeneously distributed throughout the tissue matrix, measures must be taken to negate the superchaotropicity to reinstate inter-biomolecular interactions in a
bio-orthogonal and system-wide manner. To do so, we took advantage of the enthalpy-driven chaotropic assembly reaction, where the activities of superchaotropes can be effectively negated
with supramolecular hosts in situ, reactivating interactions between the macromolecular probes and their tissue targets. Based on the above analysis, we designed a scalable deep molecular
phenotyping method, performed in two stages: a first infiltrative stage where macromolecular probes co-diffuse homogeneously with WCS with minimized reaction barriers, followed by the
addition of macrocyclic compounds for in situ host-guest reactions to reinstate antibody-antigen binding. With a much-narrowed list of chemicals to screen, we first benchmarked the
performances of several putative WCS host-guest systems using a standard protocol as previously published6,8,9 (Supplementary Fig. 4). These include perrhenate/α-cyclodextrin (ReO4−/αCD),
ferrocenium/βCD ([Fe(C5H5)2]+/βCD), _closo_-dodecaborate ions ([B12X12]2−/γCD (where X = H, Cl, Br, or I)), metallacarborane ([Co(7,8-C2B9H11)2]−/γCD), and polyoxometalates ([PM12O40]3−/γCD
(where M = Mo, or W)) (Fig. 1c, d). Group 5 and 6 halide clusters and rhenium chalcogenide clusters such as [Ta6Br12]2+, [Mo6Cl14]2- and {Re6Se8}2+ derivatives were excluded due to
instability in aqueous environments. Only ReO4-, [B12H12]2−, and [Co(7,8-C2B9H11)2]− proved compatible with immunostaining conditions without causing tissue destruction or precipitation.
[B12H12]2−/γCD produced the best staining sensitivity, specificity and signal homogeneity across depth (Supplementary Fig. 5), while the effect of derivatizing γCD was negligible
(Supplementary Fig. 5). Finally, we chose the more soluble 2-hydroxypropylated derivative (2HPγCD) for its higher water solubility in our applications. We term our method INSIHGT, for In
situ host-guest chemistry for three-dimensional histology. IN SITU HOST–GUEST CHEMISTRY FOR THREE-DIMENSIONAL HISTOLOGY (INSIHGT) INSIHGT was designed to be a minimally perturbative, deeply
and homogeneously penetrating staining method for 3D histology. Designed for affordability and scalability, INSIHGT involves simply incubating the conventional formaldehyde-fixed tissues in
[B12H12]2-/PBS with antibodies, then in 2HPγCD/PBS (Fig. 2a) - both at room temperature with no specialized equipment. We compared INSIHGT with other 3D IHC techniques using a stringent
benchmarking experiment as previously published (see “Methods”, Supplementary Fig. 4) to compare their penetration depths and homogeneity6,9. Briefly, a mouse hemibrain was first stained in
bulk for an antigen using various deep immunostaining methods (“bulk-staining”), followed by cutting the tissue coronally in the middle (thickest dimension) and re-stained for the same
marker with a different fluorophore using a standardized control method (“cut-staining”), which serves as the reference signal without penetration limitations. The tissue was then imaged on
the cut face to compare the bulk-staining intensity (deep staining method signal) and cut-staining intensity (reference signal) as a function of the bulk-staining penetration depth. We found
that INSIHGT achieved the deepest immunolabeling penetration with the best signal homogeneity throughout the penetration depth (Fig. 2b). To quantitatively compare the signal, we segmented
the labeled cells and compared the ratio between the deep immunolabelling signal and the reference signal against their penetration depths. Exponential decay curve fitting showed that the
signal homogeneity was near-ideal (Fig. 2c, Supplementary Table 1)—where there was negligible decay in deep immunolabelling signals across the penetration depth. We repeated our benchmarking
experiment with different markers, and by correlating INSIHGT signal with the reference signal, we found INSIHGT provides reliable relative quantification of cellular marker expression
levels throughout an entire mouse hemi-brain stained for 1 day (Fig. 2d). We supplemented our comparison with the binding kinetics modulating buffers employed in eFLASH and SWITCH-pumping of
mELAST tissue-hydrogel, as we lacked the specialized equipment to provide the external force fields and mechanical compressions, respectively (Supplementary Fig. 6). For SWITCH-pumping of
mELAST tissue-hydrogel, we utilized the latest protocol and buffer recipe13. Our results also showed the use of binding kinetics modulating buffers alone from eFLASH and SWITCH-pumping of
mELAST tissue-hydrogel lead to shallower staining penetration than INSIHGT, confirming the deep penetration of these methods is mainly contributed by the added external force fields and
mechanical compressions, respectively. Hence, with excellent penetration homogeneity with a simple operating protocol, INSIHGT can be the ideal method for mapping whole organs with cellular
resolution. It is also the fastest deep immunolabelling from tissue harvesting to image (Fig. 2e). Due to its compatibility with solvent-based delipidation methods, we recommend the use of
solvent-based clearing17,18,19 for an overall fastest INSIHGT protocol, although aqueous-based clearing techniques are also compatible (see “INSIHGT protocol in Supplementary Materials” for
further discussions). However, protocols involving the use of Triton X-1008,15 and triethylamine19 must be replaced with alternatives as they form precipitates with [B12H12]2−. Notably,
after washing, only a negligible effect of [B12H12]2--treatment will remain within the tissue. This is evident as the cut-staining intensity profile of INSIHGT showed very steep exponential
decay with increasing cut-staining penetration depth, and became similar to that of iDISCO (Supplementary Fig. 7) which has identical tissue pre-processing steps. Upon the addition of 2HPγCD
and washing off the so-formed complexes, the penetration enhancement effect was completely abolished. This suggests that [B12H12]2- and cyclodextrins do not further permeabilize or disrupt
the delipidated tissue. HIGH-THROUGHPUT, MULTIPLEXED, DENSE WHOLE ORGAN MAPPING After confirming INSIHGT can achieve uniform, deeply penetrating immunostaining, we next applied INSIHGT to
address the challenges in whole organ multiplexed immunostaining, where the limited penetration of macromolecular probes hinders the scale, speed, or choice of antigens that can be reliably
mapped. Due to the operational simplicity, scaling up the sample size in organ mapping experiments with INSIHGT is straightforward and can be done using multiwell cell culture plates (Fig.
3a). For example, we demonstrated our case by mapping 14 mouse kidneys in parallel (Fig. 3b) within 6 days of tissue harvesting using a standard 24-well cell culture plate. We then exemplify
the capability of INSIHGT to simultaneously map densely expressed targets in whole organs (Fig. 3c-i, Supplementary Fig. 8-9). We first performed multiplexed staining on mouse kidney with 3
days of incubation for _Lycopersicon esculentum_ lectin (LEL), Peanut agglutinin (PNA), _Griffonia simplicifolia_ lectin (GSL), and AQP-1, which are targets associated with poor probe
penetration due to their binding targets’ dense expression (Fig. 3c-d, Supplementary Fig. 2, Supplementary Fig. 9a, b). With the use of INSIHGT, the dense tubules and vascular structures can
be reliably visualized and traced (Supplementary Fig. 8). We then proceeded to map the whole brain of a 3-year-old mouse at the time of euthanasia. We utilized INSIHGT with 3 days of
staining for Calbindin (CALB1), NeuN, and c-Fos, providing cell type and activity information across the aged organ (Fig. 3e-i, Supplementary Fig. 9c). With whole organ sampling, we
identified regions where aging-related changes were prominent, these include cavitations in the bilateral thalamus and striatum (Fig. 3g, h), as well as calbindin-positive deposits in the
stratum radiatum of hippocampus (Fig. 3i). Interestingly, there seems to be an increased c-Fos expression level among the neurons surrounding thalamic cavitations (Fig. 3g) which are located
deep within the brain tissue and thus cannot be explained by preferential antibody penetration, suggesting these cavitations may affect baseline neuronal activities. Similar 1-step
multiplexed mapping of calcium-binding proteins across a whole adult mouse brain can also be performed with 3 days of staining (with a fixed tissue-to-image time of 6 days) (Fig. 3j–l,
Supplementary Movie 1). Similarly, whole adult mouse brain mapping and statistics can be obtained for ~35 million NeuN+ cells, their GABA quantities and c-Fos expression levels using the
same protocol (Supplementary Fig. 10), allowing structure, neurotransmitter, and activity markers to be analyzed simultaneously. Overall, INSIHGT overcomes technical, operational, and cost
bottlenecks towards accessible organ mapping for every basic molecular biology laboratory, providing rapid workflows to qualitatively evaluate organ-wide structural, molecular, and
functional changes in health and disease, regardless of the spatial density of the visualization target. BORON CLUSTER-BASED SUPRAMOLECULAR HISTOCHEMISTRY AS A FOUNDATION FOR SPATIAL
MULTI-OMICS With the maturation of single-cell omics technologies, integrating these high-dimensional datasets becomes problematic. Embedding these data in their native 3D spatial contexts
is the most biologically informative approach. Hence, we next tested whether our boron cluster supramolecular chemistry allows the retention and detection of multiple classes of biomolecules
and their features, based on which 3D spatial multi-omics technologies can be developed. With identical tissue processing steps and INSIHGT conditions, we tested 357 antibodies and found
323 of them (90.5%) produced the expected immunostaining patterns as manually validated with reference to the human protein atlas and/ or existing literature (Fig. 4a, Supplementary Figs.
11–15, Supplementary Table 2). This was at least six times the number of compatible antibodies demonstrated by any other deep immunostaining method (Fig. 4a), demonstrating the robustness
and scalability of INSIHGT. Antigens ranging from small molecules (e.g., neurotransmitters), epigenetic modifications, peptides to proteins and their phosphorylated forms were detectable
using INSIHGT (Fig. 4b, c). The specificity of immunostaining even allowed the degree of lysine methylations (i.e., mono-, di- and tri-methylation) and the symmetricity of arginine
dimethylations to be distinguished from one another (Fig. 4b). We further tested 21 lectins to detect complex glycosylations, proving that [B12H12]2− do not sequester divalent metal ions
essential for their carbohydrate recognition (Fig. 4d, Supplementary Fig. 16). Small molecule dyes such as nucleic acid probes, which are mostly positively charged, present a separate
challenge as they precipitate with _closo_-dodecaborates, forming [probe]n+/[B12H12]2− precipitates when co-applied with INSIHGT. We found size-matched and charge-complementing cyclodextrin
derivatives as cost-effective supramolecular host agents for non-destructive deep tissue penetration and preventing precipitation. For example, sulfobutylether-βCD (SBEβCD) (Fig. 4e) can
react with nucleic acid probes to form [probe⊂SBEβCD], which exhibits penetration enhancement during INSIHGT (Fig. 4f, g) without precipitation problems. The so-formed [probe⊂SBEβCD] complex
can thus be co-incubated with antibodies in the presence of [B12H12]2− for a simpler protocol. We also performed RNA integrity number (RIN) and whole genome DNA extraction analyses on
INSIHGT-treated samples (Supplementary Fig. 17). We found each step of the INSIHGT protocol did not result in a significant decrease in RNA integrity number (RIN) (Supplementary Fig. 17a).
The total RNA extracted after undergoing the whole INSIHGT protocol has an RIN of 7.2, compared with a RIN of 9 from a treatment-naive control sample. For whole genome DNA, both control
versus INSIHGT-protocol-treated samples have similar sample integrity and total DNA yield per mm3 sample (14.6 μg versus 10.12 μg), as well as subsequent whole genome sequencing quality
(total clean base 114.5 Gb versus 125.2 Gb) with both having a mapping rate of 99.96% (Supplementary Fig. 17b see also “Methods” on the quality control descriptions). With RNA sequencing
whole transcriptomic comparing an INSIHGT-treated sample and a paired control sample (the opposite mouse hemibrain), the results showed essentially no differentially expressed genes profiles
(Supplementary Fig. 17c). The Pearson correlation coefficient of the expression of all genes was 0.967. Hence, unsurprisingly, we found single-molecule fluorescent in situ hybridization
(FISH) is also applicable for co-detection of protein antigens and RNAs with INSIHGT. Combining all the above probes, simultaneous 3D visualization of protein antigens, RNA transcripts,
protein glycosylations, epigenetic modifications, and nuclear DNA is possible using a mixed supramolecular system in conventionally formalin-fixed intact tissue (Fig. 4h, Table 1). Taken
together, our results suggest in situ boron cluster supramolecular histochemistry can form the foundation for volumetric spatial multi-omics method development. The implication of
well-preserved RNAs suggests the possibility of post-INSIHGT section-based spatial transcriptomics. CENTIMETER-SCALE 3D HISTOCHEMISTRY BY ISOLATED DIFFUSIONAL PROPAGATION Since antibody
penetration remains the most challenging obstacle, we focus the remainder of our investigation on larger-scale 3D immunophenotyping. We thus applied INSIHGT to visualize centimeter-scale
human brain samples, without using any external force fields to drive the penetration of macromolecular probes. These large, pigmented samples were sliced in the middle of the tissues’
smallest dimensions to allow imaging of the deepest areas with tiling confocal microscopy. We show that INSIHGT can process a 1.5 cm × 1.5 cm × 3 cm human cortex block for parvalbumin (PV)
(Fig. 5a–c), with excellent homogeneity and demonstration of parvalbumin neurons predominantly in layer 4 of the human cortex. We then scaled INSIHGT to a 1.75 cm × 2.0 cm × 2.2 cm human
cerebellum block for blood vessels (using _Griffonia simplicifolia lectin I, GSL-I_) (Fig. 5d–f). As light-sheet microscopy is suboptimal due to the large human sample, we assessed the
INSIHGT staining penetration on the cut face along the thickest dimension using confocal microscopy (Fig. 5e, Supplementary Fig. 18). This again reveals excellent homogeneity with no decay
of signal across the centimeter of penetration depth. This shows that the use of boron cluster-based host-guest chemistry remains applicable for highly complex environments at the centimeter
scale. The results further show that macromolecular transport within a dense biological matrix can remain unrestricted in a non-denaturing manner by globally adjusting inter-biomolecular
interactions. We further applied INSIHGT to a 1.0 cm × 1.4 cm × 1.4 cm human brainstem with dementia with Lewy bodies (DLB) for phosphorylated alpha-synuclein at serine 129 (αSyn-pS129)
(Fig. 5g-i, Supplementary Fig. 19). The large scale of imaging enabled registration and hence correlation with mesoscale imaging modalities such as magnetic resonance imaging (MRI) (Fig. 5g,
Supplementary Movie 2). With this, we confirmed the localization of Lewy body pathologies to the locus ceruleus complex–subcerulean nuclei27 and substantia nigra, in keeping with the
prominent rapid eye movement sleep behavior disorder (RBD) symptoms of this patient. Such a radio-histopathology approach would allow for correlative structural-molecular studies for
neurodegenerative diseases. Overall, the capability of INSIHGT in achieving centimeter-sized tissue staining bridges the microscopic and mesoscopic imaging modalities, providing a general
approach to correlative magnetic resonance-molecular imaging. VOLUMETRIC SPATIAL MORPHO-PROTEOMIC CARTOGRAPHY FOR CELL TYPE IDENTIFICATION AND NEUROPEPTIDE PROXIMITY ANALYSIS We next
extended along the molecular dimension on conventionally fixed tissues, where highly multiplexed immunostaining-based molecular profiling in 3D had not been accomplished previously. A single
round of INSIHGT-based indirect immunofluorescence plus lectin histochemistry can simultaneously map up to 6 antigens (Supplementary Fig. 20), tolerating a total protein concentration at
>0.5 μg/μl in the staining buffer, and is limited only by spectral overlap and species compatibility. To achieve higher multiplexing, antibodies can be stripped off with 0.1 M sodium
sulfite in the [B12H12]2--containing buffer after overnight incubation at 37 °C (Fig. 6a, Supplementary Fig. 21). Since [B12H12]2− does not significantly disrupt intramolecular and
intermolecular noncovalent protein interactions, the approach can be directly applied to routine formaldehyde-fixed tissues, we observed no tissue damage and little distortion, obviating the
need for additional or specialist fixation methods. We exemplified this approach by mapping 28 marker expression levels in a 2 mm-thick mouse hypothalamus slice over 7 imaging rounds (Fig.
6a–c, Supplementary Figs. 22, 23). With each iterative round taking 48 h (including imaging, retrieval and elution), the whole manual process from tissue preparation to the 28-plex image
took 16 days. After registration and segmentation using Cellpose 2.028 (Fig. 6d, e, see “Methods”), we obtained 192,075 cells and their differentially expressed proteins (DEPs) based on
immunostaining signals. Note that other user-friendly approaches such as StarDist29 and BCFind30 can also be used. Omitting 3 blood vessel channels, we then obtained the normalized mean
intensities of the remaining 25 markers, their standard deviations (S.D.s) of signal intensities of the same 25 markers, as well as their distance to the nearest vessels for dimensionality
reduction analysis and clustering. The S.D.s of signal intensities for each cell served as a measure of heterogeneous expression of a certain marker within the cell (e.g., strictly
cytoplasmic or nuclear expression will have a higher S.D. than a marker expressing in both the cytoplasms and nuclei, as illustrated in Fig. 6e). Uniform manifold approximation and
projection (UMAP) analysis of a subset of 84,139 cells based on these 51 markers (Fig. 6f, Supplementary Figs. 24, 25) plus their distance to the nearest vessels revealed 42 cell type
clusters, allowing their 3D spatial interrelationships to be determined (Supplementary Fig. 26). INSIHGT allows both 3D morphology and molecular information to be well-visualized via
immunostaining, which is more difficult to access via current section-based spatial transcriptomics or single-cell multi-omics despite ongoing efforts31. Recent characterizations of neuronal
network activities based on the diffusional spread of neuropeptides highlight the need for 3D spatial mapping of protein antigens. To obtain these morphological-molecular relationships
using INSIHGT, we segmented the neuropeptide Y (NPY)-positive fibers and computed the 3D distance to each UMAP-clustered cell types’ somatic membrane (Fig. 6g–j). While most clusters have a
similar distance from NPY fibers, certain clustered cells (notably right tile clusters 1 and 2) are more proximally associated with NPY fibers, suggesting these cell clusters are
differentially modulated by NPY when isotropic diffusion is assumed in the local brain parenchyma. Nonetheless, our dataset and analysis demonstrated it is possible to estimate the likely
modulatory influence for a given cell-neuropeptide pair, providing an alternative approach to discovering neuronal dynamics paradigms. FINE-SCALE 3D IMAGING REVEALS UNSUSPECTED INTERCELLULAR
CONTACTS TRAVERSING THE BOWMAN SPACE IN MOUSE KIDNEYS We found that the process of INSIHGT from fixation to completion preserves delicate structures such as free-hanging filaments and
podia, enabling fine-scale analysis of compact structures such as the renal glomeruli. We found unsuspected intercellular contacts traversing the Bowman space, which was not known to be
present in normal glomeruli even with serial sectioning electron microscopy studies32,33,34,35,36 (Fig. 7a, b). These filaments are mostly originated from the podocytic surface, although
some were also seen to emerge from PECs. They were numerous and found around the glomerular globe (Fig. 7c), with varied in their length, distance from each other, and morphologies (Fig. 7d,
Supplementary Fig. 27). We classified these podocyte-to-PEC microfilaments into “reachers” and “stayers”, depending on whether they reached the PEC surface or not (Fig. 7e). Microfilaments
of the reachers-type were more numerous than the stayers-type per glomerulus (Fig. 7f). Visually, we noted the emergence of these filaments tended to cluster together, especially for the
reachers-type. To quantify such spatial clustering, we calculated the glomerular surface geodesic distances between the podocytic attachment points for each microfilament, which showed an
inverse relationship with their path lengths (Fig. 7g), and reachers-type filament are geodesically located nearer to each other than the stayers type (Supplementary Fig. 28). This suggests
that the emergence of long, projecting microfilaments that reach across the Bowman space is localized on a few hotspots of the glomerular surface. Whether these hotspots of long-reaching
microfilaments are driven by signals originated from the podocyte, the glomerular environment underneath, or the nearest PECs across the Bowmann space remains to be investigated and may
reveal previously unsuspected podocyte physiological responses within their microenvironments. Notably, similar structures have been observed in the pathological state of cresenteric
glomerulonephritis, in conjunction with whole cells traversing the Bowman space. As cresenteric glmoerulonephiritis is a final common pathway of glomerulonephropathies, it would be
interesting to investigate whether there is a continuum of progressive changes from microfilaments physiologically to larger trans-Bowman space connections pathologically. In addition,
morphologically similar structures have been observed in the microglia37, pericytes38, between tumor and immune cells39, and between normal and apoptotic cells in cell culture40. The
podocyte-PEC connections described here thus add another organ to the growing list of nanostructural connections mediating information and matter exchange between different cell types in
their physiological states. SPARSELY DISTRIBUTED NEUROFILAMENT INCLUSIONS UNIQUE TO THE HUMAN CEREBELLUM We next completely mapped a 3 mm-thick (post-dehydration dimensions) human cerebellar
folium for NF-H, GFAP, and blood vessels (Fig 8a, Supplementary Figs. 29, 30, Supplementary Movie 3), with preserved details down to the Bergmann glia fibers, perivascular astrocytic
endfeet, and Purkinje cell axons that make the amenable to 3D orientation analysis and visualization (Fig. 8b-d, Supplementary Figs. 29, 30). The detailed visualization of filamentous
structures throughout the 3 mm-thickness is in stark contrast to our previous attempts with similar specimens employing various methods, which showed weak NF-H signal in cerebellar sulci and
barely visible GFAP signal in cerebellar white matter due to poor antibody penetration. We discovered sparsely distributed NF-H-intense inclusions that are easily missed in 2D sectioning
and thus remain poorly characterized. We manually traced and identified 1078 inclusions throughout the entire imaged volume (Fig. 8e, f), where they were found in all of the three basic
layers of the cerebellar cortex. A typical morphology of one type of these inclusion is a single bright globular inclusion at the sub-Purkinje layer radial location, with an elongated thick
fiber extension that coils back and project to the adjacent molecular layer (Fig. 8e). However, much more protean morphologies also exist (Fig. 8e, f, Supplementary Fig. 30). To capture the
morphological and spatial diversities of these inclusions, we obtained their spatial-morphometric statistics (Supplementary Fig. 31a), followed by principal component analysis of the
compiled morphometrics such as Sholl analysis and Horton-Strahler number. The results reveal most of these inclusions to be morphologically homogeneous with variations explained largely by
their path lengths, with a small subset characterized by much higher branching of the NF-H-intense filaments (Supplementary Fig. 31b). However, further understanding of these inclusions
awaits broader investigations in normal and various disease states other than in DLB. Preliminarily, we have also observed these inclusions in normal human cerebellum tissues (Supplementary
Fig. 31c). With the advancements in technologies, correlated mulit-pronged approaches using superresolution microscopy, electron microscopy and spatially resolved proteomics are expected to
help greatly clarify the pathobiology of these inclusions. INSIHGT BRIDGES THE GAP BETWEEN 3D HISTOLOGY AND TRADITIONAL 2D PATHOLOGY IN CURRENT CLINICAL PRACTICE The bio-orthogonal nature of
the INSIHGT chemical system underlies its non-destructiveness. To highlight the clinical impact of INSIHGT in addition to 3D imaging of human samples, we found that INSIHGT-processed
samples can be retrieved and processed as naïve tissues for traditional 2D histology via paraffin wax embedding and sectioning. Notably, staining qualities of routine hematoxylin and eosin
(H&E) and various special stains on the post-INSIHGT processed slides were indistinguishable from the pre-INSIHGT processed slides even by a senior pathologist (Fig. 8g, h). In addition
to not interfering with downstream clinical processes, the preserved quality of special staining allows for multi-modal cross-validation of 3D fluorescent imaging findings, making INSIHGT
the ideal platform choice for next-generation histopathology (Fig. 8i). Together with the possibility for post-INSIHGT DNA and RNA sequencing, we envision (Supplementary Fig. 17)
quantitative 3D information within clinical specimens can be maximally extracted and preserved with high authenticity in a non-consumptive manner using INSIHGT, and its fast speed promises
compatibility with current clinical workflows and constraints, allowing digital pathology and precision medicine to benefit from 3D analysis. DISCUSSION The convergence of multiple
technological advances has paved the way for the acquisition of large-scale molecular phenotyping datasets at single-cell resolution, most notably single-cell transcriptomics36. With a large
number of previously undiscovered cell states, the quest to extend towards spatially resolved cell phenotyping based on translated and post-translationally expressed biomolecular signatures
is paramount to understanding their structural and functional properties in biology41. Scalable, high-resolution 3D tissue mapping provides a powerful approach to further our understanding
of these previously unidentified cell types. Clinically, 3D histology has been shown to improve diagnosis in bladder cancer42, predict biochemical recurrence in prostate cancer43, and
evaluate response to chemotherapy in ovarian carcinoma42, By sampling across whole intact samples, 3D histology can deliver unbiased, quantitative, ground-truth data on the spatial
distributions of molecules and cell types in their native tissue contexts44. However, 3D tissue imaging is yet to be widely adopted despite the increasing accessibility of tissue clearing,
optical sectioning microscopy, and coding-free image processing software. This is in large part due to the limited penetration of probes that plague the field regardless of the combinations
of these technologies employed6,10, yielding variable, surface-biased data with questionable representativeness. Creative approaches have provided solutions to the penetration problem but
are limited in their scalability and accessibility6. Constrained by the requirements of non-advective approaches and compatibility with off-the-shelf reagents, the development of INSIHGT
involved re-examining biomolecular transport and protein stability from the first principles, which led us to identify weakly coordinating superchaotrope and its chemical activity modulation
by in situ host–guest reactions to implement our theoretical formulation. With the use of _closo_-dodecaborate and cyclodextrin as an additive in PBS, we solved the bottleneck of 3D
histology by providing a cost-efficient, scalable, and affordable approach to quantitatively map multiple molecules in centimeter-sized tissues. With an equivalent tissue processing pipeline
to iDISCO17, INSIHGT shares the same affordability and scalability while providing much faster processing and greatly improved image quality, due to enhanced antibody penetration depth and
homogeneity. Mapping tissue blocks simultaneously in multi-well dishes is easily accomplished in any basic molecular biology laboratory. Such simplicity in operation makes it highly
accessible and automatable, as it requires no specialized equipment or skills. Furthermore, cocktails of off-the-shelf antibodies can be directly added to PBS supplemented with [B12H12]2−.
Finally, we note that both [B12H12]2− salts and cyclodextrins are non-hazardous and stable indefinitely at ambient temperatures16. With the affordability and accessibility of INSIHGT, we
anticipate its diverse applications in 2D and 3D histology applications. Meanwhile, boron cluster-based supramolecular histochemistry can form the backbone for 3D spatial
molecular-structural-functional profiling methods and studies, as well as atlas mapping efforts. The high-depth, quantitative readout of well-preserved tissue biomolecules offered by INSIHGT
forms the foundation for multiplexed, multi-modal, and multi-scale 3D spatial biology. By making non-destructive 3D tissue molecular probing accessible, INSIHGT can empower researchers to
bridge molecular-structural inferences from subcellular to the organ-wide level, even up to clinical radiological imaging scales for radio-histopathological correlations. Finally, the
compatibility of INSIHGT with downstream traditional 2D histology methods indicates its non-interference with subsequent clinical decision-making. This paves the way for the translation and
development of 3D histology-based tissue diagnostics, promising rapid and accurate generation of groundtruth data across entire tissue specimens. We recognize that INSIHGT still has room for
further improvements. Immunostaining penetration homogeneities for larger tissues and denser antigens can be further enhanced, Practically, this is limited to a maximum of ~2 cm3 sized
tissues, and extremely dense antigens such as GAPDH, type I collagen, actin, and myosin remain difficult for whole organ staining with homogeneous penetration. Nonetheless, for any antigens
stained using the iDISCO+ protocol45 with 7 days of primary antibody staining, INSIHGT with 3 days of antibody staining will at least provide 10–20× penetration enhancement, along with a
noticeable enhancement in penetration homogeneity. Penetration can be further enhanced by prolonging the incubation times and ensuring an adequate amount probes has been added relative to
the tissue expression level (see “Supplementary Note and INSIHGT protocol therein”). If available, the use of primary nanobodies with fluorescently-labeled secondary whole IgGs will further
increase the penetration by about 5–10 times. In addition, the penetration homogeneity of small molecule dyes and lectins were still suboptimal for millimeter-scale tissues and remains to be
further enhanced. In multi-round immunostaining, we noticed that the staining specificity and sensitivity deteriorated with each round of antibody elution with sulfite or β-mercaptoethanol,
calling for a better 3D immunostaining elution method. Alternatively, hyperspectral imaging46, nonlinear optics47, time-resolved fluorescence techniques48, and same-species antibody
multiplexing49 could be explored to extend the multiplexing capabilities of INSIHGT. Finally, although theoretically applicable, we have yet to apply the INSIHGT-based multi-round staining
in tissues from other species. Our discovery of boron clusters’ capabilities to solubilize proteins globally in a titratable manner, combined with their bio-orthogonal removal with
supramolecular click chemistry, can reach beyond histology applications. Given the surprisingly robust performance of INSIHGT in complex tissue environments, we envision they can be applied
in simpler in vitro settings to control intermolecular interactions—particularly when involving proteins—in a spatiotemporally precise manner. METHODS ETHICAL STATEMENT For animal tissues,
all experimental procedures were approved by the Animal Research Ethics Committee of the Chinese University of Hong Kong (CUHK) and were performed in accordance with the Guide for the Care
and Use of Laboratory Animals (AEEC number 20-287-MIS). The housing of animals was provided by the Laboratory Animal Service Center of CUHK. For human tissues donated post-mortem, prior
ethics approvals have been obtained and approved by the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (approval number 2022.137), with
consent obtained from the donor and his family. CHEMICALS AND REAGENTS The antibodies utilized in this study were listed in Supplementary Table 2. All protein-conjugating fluorophores
tested and their compatibility with INSIHGT were listed in Supplementary Table 3. Secondary Fab fragments or nanobodies were acquired from Jackson ImmunoResearch or Synaptic Systems, and all
lectins were sourced from VectorLabs. Conjugation of secondary antibodies and lectins with fluorophores was achieved through _N_-hydroxysuccinimidyl (NHS) chemistry. The process was
conducted at room temperature for a duration exceeding 16 h at antibody concentrations >3 mg/ml, using a tenfold molar excess of the reactive dye-NHS ester.
Dodecahydro-_closo_-dodecaborate salts and other boron cluster compounds were procured from Katchem, while cyclodextrin derivatives were obtained from Cyclolab, Cyclodextrin Shop, or Sigma
Aldrich. We noticed occasionally the chemicals involved in the INSIHGT process require purification. Specifically, for Na2[B12H12], if insoluble flakes were noticed after dissolution in PBS,
the solution was then acidified to pH 1 with concentrated hydrochloric acid, extracted with diethyl ether (Sigma Aldrich), and the organic solvent was removed and distilled off with a warm
water bath. The residual H2B12H12 was then dissolved in minimal amount of water, and neutralized with 1 M Na2CO3 solution until pH 7 is reached with no further evanescence. The solution was
then concentrated by distillation under vacuum and dried in an oven. For 2-hydroxypropyl-γ-cyclodextrin and sulfobutylether-β-cyclodextrin, if insoluble specks or dusts were noticed after
dissolution in PBS, the solution was vacuum filtered through 0.22μm hydrophilic cellulose membrane filters (GSWP14250) using a Buchner funnel before use. A slight brownish-yellow
discoloration of the resulting solution would not interfere with the INSIHGT results. For benzyl benzoate, if the solution is yellowish (possibly due to the impurities of fluorenone
present), the solvent is poured into a metal bowl or glass crystallization dish and refrigerated to 4 ०C until crystallization begins. If no crystallization occurs, a small crystal seed of
benzyl benzoate obtained by freezing the solvent at −20 ०C in a microcentrifuge tube can be put into the cooled solvent to kick-start the process. The crystals were then collected by vacuum
filtration with air continuously drawn at room temperature until the crystals are white, which were warmed to 37 ०C to result in clear, colorless benzyl benzoate. If the resulting colorless
benzyl benzoate is cloudy, 3 Å molecular sieves were added to the solvent to absorb the admixed water from condensation, before filtering off to result in a clear colorless benzyl benzoate.
This purified benzyl benzoate is ready to constitute BABB clearing solution for imaging. HUMAN AND ANIMAL TISSUES Adult male C57BL/6 were utilized. These mice were housed in a controlled
environment (22–23 °C) with a 12-h light-dark cycle, provided by the Laboratory Animal Service Center of CUHK. Unrestricted access to a standard mouse diet and water was ensured, and the
environment was maintained at <70% relative humidity. Tissues were perfusion formaldehyde-fixed and collected by post-mortem dissection. In the case of immunostaining for
neurotransmitters where Immusmol antibodies were used, the tissues were perfusion-fixed with the STAINperfect™ immunostaining kit A (Immusmol) with the antibody staining steps replaced with
those in our INSIHGT method. For human tissues, brain and kidney tissues donated post-mortem by a patient (aged 77 at the time of passing) were used in this study. Prior ethics approvals
have been obtained and approved by the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (approval number 2022.137), with consent from the
donor and his family. Human dissection was performed by an anatomist (HML) after perfusion fixation with 4% paraformaldehyde via the femoral artery. The post-mortem delay to fixation and
tissue harvesting was 4 weeks at −18 °C refrigeration, and the fixation duration was 1 week at room temperature. The corresponding organs were then harvested and stored in 1x PBS at room
temperature until use. SCREENING DEEP STAINING APPROACHES WITH IN SITU ANTIBODY RECOVERY 4% PFA-fixed, 1mm-thick mouse cerebellum slices, 0.5 μg anti-parvalbumin antibody (Invitrogen,
PA1-933), and 0.5 μg AlexaFluor 647-labeled Fab fragments of Donkey anti-Rabbit antibody (Jackson Immunoresearch 711-607-003) were used in this experiment to develop our method.
Co-incubation of the secondary Fab fragment and primary antibody was utilized for 1-step immunostaining. All stainings were performed with an overnight immunostaining first stage at room
temperature (unless specified otherwise) in various buffers, with subsequent recovery secondary stage at room temperature (unless specified otherwise) in various buffers, as detailed for
each strategy below. The tissues were then washed in 1x PBSN, dehydrated with graded methanol, and cleared in BABB, before proceeding to imaging with confocal microscopy. For the SDS/αCD
system, immunostaining was performed in a solution consisting of 10 mM sodium dodecylsulphate (SDS) in 1xPBS, while recovery was performed with a solution consisting of 10 mM αCD in 1x PBS.
For the GnCl/GroEL+GroES system, immunostaining was performed in solution consisting of 6 M guanidinium chloride in 1x PBS, while recovery was performed with GroEL+GroES refolding buffer,
consisting of 0.5 μM GroEL (MCLabs GEL-100), 1 μM GroES (MCLabs GES-100), 2.5 mM adenosine triphosphate, 20 mM Tris base, 300 mM NaCl, 10 mM MgSO4, 10 mM KCl, 1 mM
tris(2-carboxylethyl)phosphine hydrochloride, 10% glycerol, with pH adjusted to 7.950. For the sodium deoxycholate (SDC)/βCD system, immunostaining was performed in a solution consisting of
15 mM sodium deoxycholate (SDC) with 240 mM Tris base, 360 mM CAPS (_N_-cyclohexyl-3-aminopropanesulfonic acid), with pH adjusted to 8, while recovery was performed with a solution
consisting of 15 mM βCD with 240 mM Tris base, 360 mM CAPS, with pH adjusted to 8. For the Na2[B12H12]/γCD system, immunostaining was performed in a solution consisting of 0.1 M Na2[B12H12]
in 1x PBS, while recovery was performed in a solution consisting of 0.1 M γCD in 1x PBS. BENCHMARKING EXPERIMENTS We designed a stringent benchmarking scheme for quantitative evaluation of
antibody penetration depth and signal homogeneity across depth for comparison across existing deep immunostaining methods, based on our previously described principles (Supplementary Fig.
1a)6 The benchmarking experiment is carried out in two parts, the first part using a whole mouse hemisphere stained in bulk with anti-Parvalbumin (PV) antibodies with excess AlexaFluor
647-conjugated secondary Fab fragments—termed bulk-staining—after which the tissue is cut coronally at defined locations using a brain matrix and re-stained with anti-PV antibodies and
AlexaFluor 488-conjugated secondary Fab fragments—termed cut-staining (Supplementary Fig. 1a). Hence, signals from bulk-staining can be distinguished easily from cut-staining and reveal
different penetration depths of the two-staged immunostaining. We tested different deep immunostaining methods in the bulk-staining stage of the experiments, while the cut-staining was
performed in 1× PBS with 0.1% Tween-20 as a conventional immunostaining buffer. The bulk-staining duration for INSIHGT was 24 h in benchmarking. All benchmarking samples were perfusion-fixed
with 4% paraformaldehyde (PFA) in 1× PBS followed by post-fixation in 4% PFA overnight at 4 °C, except for SHIELD and mELAST samples where the SHIELD protocol was used. In addition, the
final RI matching where the benzyl alcohol/benzyl benzoate (BABB) clearing method was universally employed to standardize the changes in tissue volumes and hence penetration distance
adjustments. The standardized optical clearing avoids the variability in fluorescent quenching and tissue shrinkage/expansion introduced by different RI matching agents. For bulk-staining
during our benchmarking experiment, we followed the published protocols except for eFLASH and mELAST due to the lack of specialized in-house equipment. For eFLASH12, we stained the SHIELDed
and SDS-delipidated tissue in the alkaline sodium deoxycholate buffer (240 mM Tris, 160 mM CAPS, 20% w/v D-sorbitol, 0.9% w/v sodium deoxycholate) and titrated-in acid-adjusting booster
buffer (20% w/v D-sorbitol and 60 mM boric acid) hourly over 24 h to achieve a −0.1 ± 0.1 pH/h adjustment rate, using primary IgGs with secondary fluorophore-labeled Fab fragments. The
tissue was then washed with 1× PBSTN (1× PBS, 1% v/v Triton X-100, and 0.02% w/v NaN3) two times 3 h each before imaging. For mELAST7,13,14, we stained the SHIELDed and SDS-delipidated
tissue with the antibody and Fab fragments in 0.2 × PBSNaCh (0.2× PBS, 5% w/v NaCh and 0.02% w/v NaN3, 5% v/v normal donkey serum) first for 1 day at 37 °C without embedding the SHIELDed
tissue in elastic gel nor compression/stretching, followed by adding Triton X-100 to a final concentration of ~5% and incubated for 1 more day. The tissue was then washed with 1× PBSTN 2
times 3 h each before imaging. For CUBIC HistoVision8 and iDISCO17, the tissue was processed and stained as previously described9. The staining durations were 14 days for CUBIC HistoVision
and 7 days for iDISCO (both using primary IgGs with secondary fluorophore-labeled Fab fragments). For SHANEL51, the tissue was first delipidated with CHAPS/NMDEA solution (10% w/v CHAPS
detergent and 25% w/v _N_-methyldiethanolamine in water) for 1 week, then further delipidated with dichloromethane/methanol as in iDISCO, then treated with 0.5 M acetic acid for 2 days,
washed in water for 6 h repeated 2 times, and then treated with guanidinium solution (PBS with 4 M guanidinium chloride, 0.05 sodium acetate, 2% w/v Triton X-100) for 2 days, blocked in
blocking buffer (1× PBS, 0.2% v/v Triton X-100, 10% v/v DMSO, 10% goat serum) for 1 day, and finally stained in antibody incubation buffer (1× PBS, 0.2% v/v Tween-20, 3% v/v DMSO, 3% v/v
goat serum, 10 mg/L heparin sodium) using primary IgGs with secondary fluorophore-labeled Fab fragments for 7 days. For quantification, PV-positive cells were identified using a Laplacian of
Gaussian filter, followed by intensity-based segmentation. These segmented masks allow the quantification of bulk- and cut-staining channel intensities, in addition to the distance
transformation intensity, performed in MATLAB R2023a (MathWorks, US). For an ideal deep immunostaining, the bulk-immunostaining signals should be independent of the bulk-staining penetration
distances computed with distance transform of the segmented tissue boundaries, and perfectly correlate with that of cut-immunostaining. This is often not the case, as “rimming” of
bulk-staining signals inevitably occurs as a “shell” around the tissue due to more easily accessible antigens on the bulk-staining tissue surface. The rimming effect can be quantified by
fitting a single-term exponential decay curve $$\frac{{{{\rm{bulk-staining}}}}\; {{{\rm{intensity}}}}}{{{{\rm{cut-staining}}}}\; {{{\rm{intensity}}}}}={e}^{-\tau ({{{\rm{bulk-staining}}}}\;
{{{\rm{penetration}}}}\; {{{\rm{distance}}}})}$$ (1) and evaluating the decay constant, tau (τ), across penetration depths, with τ → 0+ as we approach the ideal case. SCREENING CHEMICALS FOR
INSIHGT We first pre-screened the WCS by immunostaining for parvalbumin in 1 mm3 of mouse cortex tissue cubes in the presence of WCS at 0.1 M, after 1 day of incubation at room temperature
the staining solution was aspirated and 0.1 M corresponding cyclodextrin was added and incubated overnight. The tissue was then washed in PBSN for 15 min two times and cleared with the BABB
method, and imaged. This procedure eliminated [B12Br12]2−, [B12I12]2−, and [PW12O40]3− (as cesium or sodium salts) as they do not give the correct immunostaining pattern or lead to tissue
destruction. We tested [Fe(C5H5)2]+ (as the hexafluorophosphate salt) for the sake of completion as a low-charge large-sized cation. To benchmark the ability in achieving deep and
homogeneous immunostaining, the above benchmarking procedure was used. Mouse hemibrains were fixed, washed, and stained with 1 μg rabbit anti-parvalbumin antibody with 1 μg AlexaFluor
647-labeled donkey anti-rabbit secondary antibody Fab fragments in 0.1 M of the WCS. The staining proceeded for 1 day after which the solution was replaced with 0.1 M corresponding
cyclodextrin (or its derivatives) and incubated overnight. The hemibrains were then washed in PBSN for 1 h two times, cut in the middle coronally and re-stained for parvalbumin using
AlexaFluor 488-labeled secondary Fab fragments. The tissue was then washed, cleared with the BABB method, and imaged on the cut face using a confocal microscope. INSIHGT A detailed
step-by-step protocol used in this study has been given below. As a general overview, tissues were typically fixed using formalin or 4% paraformaldehyde, thoroughly washed in PBSN, and
pre-incubated overnight at 37 °C in INSIHGT buffer A. The tissues were then stained with a solution containing the desired antibodies, Fab fragments, lectins, and SBEβCD-complexed nucleic
acid probes in INSIHGT buffer A, ensuring a final [B12H12]2− concentration of 0.25 M. Staining duration varied from 6 h to 10 days based on tissue size, antigen, and required homogeneity
(please see the calculation of time _t_ in the step-by-step protocol). Post-staining, the solution was aspirated and replaced with INSIHGT buffer B (0.25 M 2-hydroxypropyl-γ-cyclodextrin in
PBS) without prior washing, followed by a minimum 6-h incubation with adequate shaking of the viscous buffer. After sufficient PBSN washing, tissues were ready for imaging or clearing. Over
incubation for any steps up to 60 days was tolerable. After imaging, the antibodies can be eluted with 0.1 M sodium sulphite in INSIHGT buffer A at 37 °C overnight. SCREENING ANTIBODIES
COMPATIBLE WITH INSIHGT To test antibodies in a high-throughput manner, we compiled a list of antibodies, reviewed their tissue expression and staining patterns in the literature, and then
obtained the respective tissues known to have positive staining. These tissue blocks or entire organs were then washed, dehydrated, delipidated, rehydrated, washed, and infiltrated with
INSIHGT solution A as described in the INSIHGT protocol. These INSIHGT-infiltrated tissues were then cut into ~1 mm3 tissue cubes and placed in a 96-well plate as indicated in the list, with
each well containing 70 μl of 1x INSIHGT solution A. About 0.5 μg of the primary antibody to be tested was then added and 0.5 μg of the corresponding AlexaFluor 647 or AlexaFluor
594-conjugated secondary antibody Fab fragment. The AlexaFluor 647 and 594 fluorophores were chosen for to minimize interference from any tissue autofluorescence on the result
interpretation. For a total volume and antibodies added two each well, an equal volume of 2x INSIHGT solution A was then added to ensure the final concentration of 1x INSIHGT solution A. The
plate was then sealed and the staining was allowed to proceed in the dark overnight at room temperature. The tissues were then washed in INSIHGT solution B for 2 h, PBSN for 1 h for two
times, and then dehydrated with through 15 min-incubation of 50% methanol, 100% methanol, and 100% methanol. The tissues were then cleared in BABB for 15 min and proceeded to imaging. The
total fixed tissue-to-image time for the antibody compatibility test is <36 h. COMPARISON BETWEEN 2D HISTOLOGICAL STAINING OF POST-INSIHGT AND CONTROL TISSUES Mouse and human samples were
pre-processed as described above. Tissues were divided into the post-INSIHGT treated group which underwent the INSIHGT protocol with 3 days of INSIHGT A incubation without the application
of antibodies and 6 h of INSIHGT B incubation, plus BABB clearing, and the control group which was immersed in PBSN for an equivalent period of time. Both groups were immersed in 70%
ethanol, preceded by the immersion in 100% ethanol for the post-INSIHGT group (which were in BABB), and in 50% ethanol for the control group (which were in PBSN). Tissues were then immersed
in 100% ethanol, xylene, and paraffin as in the standard paraffin embedding process. The embedded tissues were cut into 5 μm (human) or 10 μm (mouse) sections followed 2D histological
staining with special stains. Following standard protocols, H&E staining was performed on human brain and kidney, PAS staining was performed on human kidney, Alcian blue staining was
performed on mouse colon, and Masson trichrome staining was performed on mouse kidney samples. MICROSCOPY Confocal microscopy was performed using a Leica SP8 confocal microscope equipped
with excitation lasers at 405 nm, 488 nm, 514 nm, 561 nm, 649 nm, with detection using a 10× (NA 0.4, Leica HC PL APO ×10/0.40 CS2) or a 40× oil-immersion (NA 1.30, Leica HC PL APO 40×/1.30
Oil CS2) objective and a tunable emission filter. A custom-built MesoSPIM v5.152 was used for light-sheet microscopy equipped with lasers at 405 nm, 488 nm, 514 nm, 561 nm, 633 nm, and 675
nm, with detection using an Olympus MVX-ZB10 zoom body with a magnification range from 0.63×–6.3×. The equipped emission filters were from AHF, including QuadLine Rejectionband
ZET405/488/561/640, 440/50 ET Bandpass, 509/22 Brightline HC, 515/LP Brightline HC Longpass Filter, 542/27 BrightLine HC, 585/40 ET Bandpass, 594 LP Edge Basic Longpass Filter, 660/13
BrightLine HC, 633 LP Edge Basic Longpass Filter, and a 685/LP BrightLine HC Longpass Filter. Two-photon tomography was performed at 780 nm excitation9 using a 16× objective (NA 0.8, Nikon
CFI75 LWD 16X W), equipped with four emission filters (ThorLabs 460-60, Semrock 525/50, Semrock 607/70, and Chroma ET 670/50). Basic image acquisition parameters for all microscopy images in
this study were listed in Supplementary Table 4. RNA AND DNA QUALITY CONTROL Control and INSIHGT-treated samples following the 1 mm3 treatment timeline were re-embedded in paraffin wax and
sent for nucleic acid integrity, sequencing, and bioinformatics analysis services provided by the BGI Hongkong Tech Solution NGS Lab. RNA integrity number analysis was performed using the
Qubit Fluorometer. Whole genome DNA quality analysis was performed using the Agilent 2100 Bioanalyzer system. Sequencing was performed using the DNBSEQTM sequencing technology platform. For
transcriptomic comparison, the total clean bases were 11.2 Gb and 10.97 Gb for the control and INSIHGT-treated samples, respectively. The clean reads ratio after filtering was 90.64% and
89.96%, respectively. For whole genome sequencing, The total clean bases were 114.5 Gb and 125.2 Gb for the control and INSIHGT-treated samples, respectively, with both samples having a
clean data rate of 100% and a mapping rate of 99.96%. RNA FISH HCR WITH INSIHGT Our RNA FISH HCR protocol is largely adapted from Choi et al.53. The post-INSIHGT samples were first fixed in
4% PFA for 1 day. The samples were then pre-incubated in pre-hybridization buffer until the tissue sank to the bottom, and hybridized in hybridization buffer at 37 °C overnight. The next
day, the tissue was washed in probe wash buffer for 1 h two times at room temperature, pre-incubated in amplification buffer for 30 min, followed by HCR amplification by incubating in
amplification buffer with the addition of 30 pmol of fluorescently-labeled HCR hairpins and incubated overnight at RT. Note that the HCR hairpins were snap-cooled (heated at 95 °C for 2 min
and cooled to RT for 30 min) in 10 μL 5× SSC buffer before application to ensure hairpin structures are formed54. The samples were then washed thoroughly in 500 μL probe wash buffer for 30
min × 3 times to mitigate non-specific binding and later subjected to confocal imaging. The HCR probes which hybridize on the mRNA targets were custom-designed following the approach by Choi
et al.53, as shown in Table 1, and were purchased from Integrated DNA Technologies. IMAGE PROCESSING No penetration-related attenuation intensity adjustments were performed for all
displayed images except for the 3D renderings (but not 2D cross-sectional views) in Fig. 3 and Supplementary Movie 1 to provide the best visualization of an internal signal. For samples
imaged with two-photon tomography, we noticed a thin rim attributed to the heat produced during the gelatin embedding process (which we verified by repeating the staining and confirming its
absence with light sheet microscopy). We employed an intensity transformation mask based on the exponent of the distance from the whole organ mask surface. Image segmentation was performed
with Cellpose 2.028 for cells implemented in MATLAB R2023b or Python, or with simple intensity thresholding. Affine and non-linear image registration was performed in MATLAB R2023a or
manually in Adobe After Effects 2020 using the mesh warp effect and time remapping for _z_-plane adjustment. Image stitching was performed either with ImageJ BigStitcher plugin55 or assisted
manually with Adobe After Effects 2020 followed by tile allocation using custom-written scripts in MATLAB R2023a. 3D image visualization and Movie rendering were performed with Bitplane
Imaris 9.1, which were done as raw data with brightness and contrast adjustments, except for the whole mouse brain imaged with two-photon tomography. To remove their slicing artifacts, we
resliced the volume into x-z slices, performed a z-direction Gaussian blur, followed by a 2D Fourier transform and filtered out non-central frequency peaks before inverting the transform.
Finally, a Richardson-Lucy deconvolution was performed with a point-spread function elongated in the x-z direction, and resliced back into x-y slices. SEGMENTATION AND ANALYSIS OF
PODOCYTE-TO-PEC MICROFILAMENTS IN MOUSE KIDNEYS Podocyte-to-PEC microfilaments of 14 mouse kidneys were manually traced via the SNT plugin in ImageJ56. Path properties of the tracings were
then exported for further analysis using custom codes in MATLAB R2023a. Distance transforms were performed under manually curated glomerulus and Bowman space masks, such that each voxel
value corresponds to the distance between that voxel and the nearest nonzero voxel of the Bowman space mask. Path displacement \({d}_{{fil}}\) was computed via Pythagoras theorem using the
start and end coordinates of the filament. Minimal distance \({d}_{\min }\) is defined as the voxel value difference between the start and end coordinates. Path length \({d}_{{path}}\) is
directly measured via SNT. Tortuosity is defined as \({d}_{{path}}/{d}_{{fil}}\), skewness is defined as \({d}_{{fil}}/{d}_{\min }\), and the angle of take-off is defined as the angle
between the unit gradient vector of the distance transform and the unit path displacement vector. The geodesic distance \({d}_{A}\left(p,q\right)\) between voxels \(p,q\in A\) is defined as
the minimal of length _L_ of path(s) _P_ = (_p_1, _p_2, …, _p_l) connecting _p_, _q_, where _A_ is the set of all voxels constituting the surface of the glomerular mask57:
$${d}_{A}\left(p,q\right)=\min \{L\left(P\right):{p}_{1}=p,{p}_{l}=q,P\subseteq A\}$$ (2) Correlation statistics were then performed via GraphPad Prism version 8 for Windows, GraphPad
Software, Boston, Massachusetts USA, www.graphpad.com. Tracing and statistical analysis for the human cerebellar neurofilament inclusions were performed analogously. SPATIAL ORIENTATION AND
FRACTIONAL ANISOTROPY VISUALIZATION OF HUMAN CEREBELLUM NEURAL AND GLIAL FILAMENTS To visualize cerebellar neural and glial fibers in their preferred orientations, we performed structure
tensor analysis with orientation-based color-coding in 3D. In detail, let \(G:{{\mathbb{R}}}^{3}\times {{\mathbb{R}}}_{+}{\mathbb{\to }}{\mathbb{R}}\) be a 3D Gaussian kernel with standard
deviation \(\sigma\): $$G\left(x,y,z,\sigma \right)=\frac{1}{{\left(2\pi {\sigma }^{2}\right)}^{\frac{3}{2}}}\exp \left(-\frac{{x}^{2}+{y}^{2}+{z}^{2}}{2{\sigma }^{2}}\right)$$ (3) Define a
3D image as a function \(I:{{\mathbb{R}}}^{{\mathbb{3}}}{\mathbb{\to }}{\mathbb{R}}\) which outputs the spatial voxel values. The gradient \({{{\boldsymbol{\nabla
}}}}{{{\bf{I}}}}:\,{{\mathbb{R}}}^{3}\to {{\mathbb{R}}}^{3}\) of \(I\) at each voxel is obtained by convolving \(I\) with the spatial derivatives of \(G\): $${{{\boldsymbol{\nabla
}}}}{{{\bf{G}}}}\left(x,y,z,\sigma \right)=\left(\frac{\partial G}{\partial x},\frac{\partial G}{\partial y},\frac{\partial G}{\partial z}\right)$$ (4) $${{{\boldsymbol{\nabla
}}}}{{{\bf{I}}}}=I * {{{\boldsymbol{\nabla }}}}{{{\bf{G}}}}$$ (5) where \(*\) denotes the convolution operation. Compute the structure tensor \({{{\rm{T}}}}:{{\mathbb{R}}}^{3}\to
{{\mathbb{R}}}^{3\times 3}\) as the outer product of \({{{\boldsymbol{\nabla }}}}{{{\bf{I}}}}\) with itself: $${{{\bf{T}}}}\left(x,y,z\right)={{{\boldsymbol{\nabla }}}}{{{\bf{I}}}}\otimes
{{{\boldsymbol{\nabla }}}}{{{\bf{I}}}}$$ (6) \({{{\bf{T}}}}\) is then smoothed over a neighborhood \(N\) via convolution with \(G\) to give \(\bar{{{{\bf{T}}}}}\):
$$\bar{{{{\bf{T}}}}}\left(x,y,z\right)=G * {{{\bf{T}}}}\left(x,y,z\right)$$ (7) $$\bar{{{{\bf{T}}}}}\left(x,y,z\right)=\left[\begin{array}{ccc}{\left\langle {I}_{x}^{2}\right\rangle }_{N}
& {\left\langle {I}_{x}{I}_{y}\right\rangle }_{N} & {\left\langle {I}_{x}{I}_{z}\right\rangle }_{N}\\ {\left\langle {I}_{y}{I}_{x}\right\rangle }_{N} & {\left\langle
{I}_{y}^{2}\right\rangle }_{N} & {\left\langle {I}_{y}{I}_{z}\right\rangle }_{N}\\ {\left\langle {I}_{z}{I}_{x}\right\rangle }_{N} & {\left\langle {I}_{z}{I}_{y}\right\rangle }_{N}
& {\left\langle {I}_{z}^{2}\right\rangle }_{N}\end{array}\right]$$ (8) where \({\left\langle \cdot \right\rangle }_{N}\) represents the Gaussian-weighted smoothing over \(N\)58,59.
Eigendecomposition of \(\bar{{{{\bf{T}}}}}\) is then performed to define the shape (eigenvalues, \(\lambda\)) and the orientation (eigenvectors, \({{{{\bf{v}}}}}_{{{{\bf{e}}}}}\)) of the
diffusion ellipsoid. The fractional anisotropy (\({FA}\)) is then computed from \(\lambda\): $${FA}=\sqrt{\frac{{\left({\lambda }_{1}-{\lambda }_{2}\right)}^{2}+{\left({\lambda
}_{2}-{\lambda }_{3}\right)}^{2}+{\left({\lambda }_{3}-{\lambda }_{1}\right)}^{2}}{2\left({\lambda }_{1}^{2}+{\lambda }_{2}^{2}+{\lambda }_{3}^{2}\right)}}$$ (9) where \({FA}\) ranges from 0
(complete isotropic diffusion) to 1 (complete anisotropic diffusion)60. The tertiary (least) eigenvalue-associated eigenvectors were then extracted for the 3-dimensional image volume, with
the 4th dimension encoding the corresponding vector basis magnitudes. To visualize the orientation of fibers in the context of the image, the eigenvectors were intensity-modulated with both
the fractional anisotropy and the original image voxel values, and represented as a 3D RGB stack for visualization in Imaris. MULTI-ROUND MULTIPLEXED 3D IMAGE PROCESSING AND ANALYSIS As the
images were acquired across multiple rounds on a confocal microscope, we encountered the issues of misalignment and z-step glitching due to piezoelectric motor errors. Hence, the tiles of
images can neither be directly stitched nor registered across multiple rounds. A custom MATLAB code was written to manually remove all the z-step glitching, followed by matching the z-steps
across multiple rounds aiding by using the time-remapping function in Adobe After Effects, with linear interpolation for the transformed z-substacks. The resulting glitch-removed, z-matched
tiles were then rigid registered using the image registration application in MATLAB, followed by non-rigid registration for local matching. Finally, the registrated tiles were stitched for
downstream processing. Before segmentation, all non-vessel channels underwent background subtraction. They were then summed to capture the full morphology of stained cells, followed by
segmentation using Cellpose 2.028. A custom model was trained and used based on 2D excerpts of the images until adequate segmentation accuracy was achieved by manual inspection. The final
test image segmentation has a Dice Coefficient (or F1-score) of 0.9354 ± 0.0596 and Jaccard Index of 0.8824 ± 0.1023, provided as mean ± S.D. on six excerpted test images. Vessels were
segmented based on their staining intensity, and a distance transform was used to obtain the distance from vessels for all voxels. The cell masks subsequently facilitated the acquisition of
the statistics for all stained channels. UMAP was performed in MATLAB R2023a using the UMAP 4.455,61 package in a nested manner, incorporating the means and standard deviations of all
immunostaining intensities, as well as the distance to the nearest blood vessel. An initial UMAP (with “min_dist” = 0.05, “metric” = “euclidean”, and “n_neighbors” = 15) was applied to each
image stack tile, followed by DBSCAN clustering (using the default value _ε_ = 0.6) to eliminate the largest cluster based on cell count. The remaining cells were subjected to a second UMAP
(with the same parameters), where another round of DBSCAN clustering (with the same parameters) yielded the final cell clusters for analysis. The choice of UMAP parameters was based on an
online guide (https://umap-learn.readthedocs.io/en/latest/api.html) and visual inspection on the reasonable clustering results. Violin plots for each clustered cell type’s distance from
neuropeptide Y-positive fibers were obtained by creating a distance transformation field from the segmented fibers. Segmented cell masks were used to compute the mean intensity value of the
distance transformation field. The pairwise distances of the clustered cell types were obtained for the 30 nearest neighbors, followed by calculating the mean and SD for the coefficient of
variation. The gramm package in MATLAB R2023a was used for plotting some of the graphs62. STATISTICS AND REPRODUCIBILITY For Fig. 2c, Supplementary Figs. 6, 7, one-component exponential
regression was applied for curve fitting, and Pearson’s correlation coefficient was computed for the scattered plot in Fig. 2d. Two-sample unpaired _t_-test was employed for Supp. Fig. 28
The staining and imaging experiments in Fig. 2–8, were repeated with at least two independent samples in the same or similar condition with slight modifications, such as using similarly
sized tissues of similar characteristics (especially for human samples), using different staining antibodies and marker choice, or staining durations. All the results were reliably
reproduced in accordance with the expected outcome of the methods. No method was used to predetermine sample size. REPORTING SUMMARY Further information on research design is available in
the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY The raw imaging data in this paper are too large for public deposit. They will be made available upon request
to the corresponding author (H.M.L). The benchmarking experiment dataset has been deposited and made available for analysis at Code Ocean (capsule link:
https://doi.org/10.24433/CO.4249201.v1). The data associated with Fig. 7f, g were provided in the Source Data File. Source data are provided with this paper. CODE AVAILABILITY The code for
benchmarking experiment analysis along with sample data has been deposited and made available at Code Ocean (capsule link: https://doi.org/10.24433/CO.4249201.v1). REFERENCES * Moses, L.
& Pachter, L. Museum of spatial transcriptomics. _Nat. Methods_ 19, 534–546 (2022). Article CAS PubMed MATH Google Scholar * Hickey, J. W. et al. Spatial mapping of protein
composition and tissue organization: a primer for multiplexed antibody-based imaging. _Nat. Methods_ 19, 284–295 (2021). Article PubMed PubMed Central MATH Google Scholar * Cho, W.,
Kim, S. & Park, Y.-G. Towards multiplexed immunofluorescence of 3D tissues. _Mol. Brain_ 16, 37 (2023). Article PubMed PubMed Central MATH Google Scholar * Liu, J. T. C., Glaser, A.
K., Poudel, C. & Vaughan, J. C. Nondestructive 3D pathology with light-sheet fluorescence microscopy for translational research and clinical assays. _Annu. Rev. Anal. Chem._ 16, 231–252
(2023). Article CAS MATH Google Scholar * Richardson, D. S. et al. Tissue clearing. _Nat. Rev. Methods Prim._ 1, 1–24 (2021). CAS MATH Google Scholar * Yau, C. N. et al. Principles
of deep immunohistochemistry for 3D histology. _Cell Rep. Methods_ 3, 100458 (2023). Article CAS PubMed PubMed Central MATH Google Scholar * Ku, T. et al. Elasticizing tissues for
reversible shape transformation and accelerated molecular labeling. _Nat. Methods_ 17, 609–613 (2020). Article CAS PubMed PubMed Central MATH Google Scholar * Susaki, E. A. et al.
Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. _Nat. Commun._ 11, 1982 (2020). Article CAS PubMed PubMed Central MATH ADS
Google Scholar * Lai, H. M. et al. Antibody stabilization for thermally accelerated deep immunostaining. _Nat. Methods_ 19, 1137–1146 (2022). Article CAS PubMed PubMed Central MATH
Google Scholar * Mai, H. et al. Whole-body cellular mapping in mouse using standard IgG antibodies. _Nat. Biotechnol_. https://doi.org/10.1038/s41587-023-01846-0 (2023). * Kim, S.-Y. et al.
Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. _Proc. Natl Acad. Sci._ 112, E6274–E6283 (2015). Article CAS PubMed PubMed Central
Google Scholar * Yun, D. H. et al. Ultrafast immunostaining of organ-scale tissues for scalable proteomic phenotyping. _bioRxiv_ 660373 https://doi.org/10.1101/660373 (2019). * Park, J. et
al. Integrated platform for multi-scale molecular imaging and phenotyping of the human brain. _bioRxiv_ 2022.03.13.484171 https://doi.org/10.1101/2022.03.13.484171 (2023). * Murray, E. et
al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. _Cell_ 163, 1500–1514 (2015). Article CAS PubMed PubMed Central MATH Google Scholar * Park,
Y.-G. et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. _Nat. Biotechnol._ 37, 73–83 (2018). Article MATH Google Scholar * Pitochelli, A. R. &
Hawthorne, F. M. The isolation of the icosahedral B12H12-2 ION. _J. Am. Chem. Soc._ 82, 3228–3229 (1960). Article CAS MATH Google Scholar * Renier, N. et al. iDISCO: a simple, rapid
method to immunolabel large tissue samples for volume imaging. _Cell_ 159, 896–910 (2014). Article CAS PubMed MATH Google Scholar * Dent, J. A., Polson, A. G. & Klymkowsky, M. W. A
whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. _Development_ 105, 61–74 (1989). Article CAS PubMed Google Scholar *
Masselink, W. et al. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. _Development_ 146, dev166884 (2019). * Scardigli, M. et al. Comparison of different tissue
clearing methods for three-dimensional reconstruction of human brain cellular anatomy using advanced imaging techniques. _Front. Neuroanat._ 15, 752234 (2021). Article CAS PubMed PubMed
Central MATH Google Scholar * Darche, M. et al. Light sheet fluorescence microscopy of cleared human eyes. _Commun. Biol._ 6, 1–7 (2023). Article Google Scholar * Cartmell, S. C. et al.
Multimodal characterization of the human nucleus accumbens. _Neuroimage_ 198, 137–149 (2019). Article PubMed MATH Google Scholar * Lichtenegger, A. et al. Assessment of pathological
features in Alzheimer’s disease brain tissue with a large field-of-view visible-light optical coherence microscope. _Neurophotonics_ 5, 035002 (2018). Article PubMed PubMed Central MATH
Google Scholar * Gail Canter, R. et al. 3D mapping reveals network-specific amyloid progression and subcortical susceptibility in mice. _Commun. Biol._ 2, 360 (2019). Article PubMed
PubMed Central Google Scholar * Salvi, G., De Los Rios, P. & Vendruscolo, M. Effective interactions between chaotropic agents and proteins. _Proteins_ 61, 492–499 (2005). Article CAS
PubMed MATH Google Scholar * Proc, J. L. et al. A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins
by trypsin. _J. Proteome Res._ 9, 5422–5437 (2010). Article CAS PubMed PubMed Central MATH Google Scholar * Dugger, B. N. et al. Neuropathological analysis of brainstem cholinergic and
catecholaminergic nuclei in relation to rapid eye movement (REM) sleep behaviour disorder. _Neuropathol. Appl. Neurobiol._ 38, 142–152 (2012). Article CAS PubMed PubMed Central MATH
Google Scholar * Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. _Nat. Methods_ 19, 1634–1641 (2022). Article CAS PubMed PubMed Central MATH Google Scholar
* Weigert, M., Schmidt, U., Haase, R., Sugawara, K. & Myers, E. Star-convex polyhedra for 3D object detection and segmentation in microscopy. In _Proc. IEEE Workshop on Applications of
Computer Vision_ 3655–3662 (IEEE, 2019). * Frasconi, P. et al. Large-scale automated identification of mouse brain cells in confocal light sheet microscopy images. _Bioinformatics_ 30,
i587–i593 (2014). Article CAS PubMed PubMed Central MATH Google Scholar * Bao, F. et al. Integrative spatial analysis of cell morphologies and transcriptional states with MUSE. _Nat.
Biotechnol._ 40, 1200–1209 (2022). Article CAS PubMed MATH Google Scholar * Shankland, S. J., Smeets, B., Pippin, J. W. & Moeller, M. J. The emergence of the glomerular parietal
epithelial cell. _Nat. Rev. Nephrol._ 10, 158–173 (2014). Article CAS PubMed Google Scholar * Li, Z.-H. et al. The role of parietal epithelial cells in the pathogenesis of podocytopathy.
_Front. Physiol._ 13, 832772 (2022). Article PubMed PubMed Central Google Scholar * Neal, C. R. et al. Novel hemodynamic structures in the human glomerulus. _Am. J. Physiol. Ren.
Physiol._ 315, F1370–F1384 (2018). Article MATH Google Scholar * Ohse, T. et al. The enigmatic parietal epithelial cell is finally getting noticed: a review. _Kidney Int._ 76, 1225–1238
(2009). Article PubMed PubMed Central MATH Google Scholar * Potter, S. S. Single-cell RNA sequencing for the study of development, physiology and disease. _Nat. Rev. Nephrol._ 14,
479–492 (2018). Article CAS PubMed PubMed Central MATH Google Scholar * Chakraborty, R., Nonaka, T., Hasegawa, M. & Zurzolo, C. Tunnelling nanotubes between neuronal and microglial
cells allow bi-directional transfer of α-Synuclein and mitochondria. _Cell Death Dis._ 14, 329 (2023). Article CAS PubMed PubMed Central MATH Google Scholar * Alarcon-Martinez, L. et
al. Interpericyte tunnelling nanotubes regulate neurovascular coupling. _Nature_ 585, 91–95 (2020). Article CAS PubMed ADS Google Scholar * Saha, T. et al. Intercellular nanotubes
mediate mitochondrial trafficking between cancer and immune cells. _Nat. Nanotechnol._ 17, 98–106 (2022). Article CAS PubMed MATH ADS Google Scholar * Wang, X. & Gerdes, H.-H.
Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. _Cell Death Differ._ 22, 1181–1191 (2015). Article CAS PubMed PubMed Central MATH Google Scholar *
Piwecka, M., Rajewsky, N. & Rybak-Wolf, A. Single-cell and spatial transcriptomics: deciphering brain complexity in health and disease. _Nat. Rev. Neurol._ 19, 346–362 (2023). Article
PubMed PubMed Central Google Scholar * Tanaka, N. et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. _Nat. Biomed. Eng._ 1,
796–806 (2017). Article CAS PubMed MATH Google Scholar * Xie, W. et al. Prostate cancer risk stratification via nondestructive 3d pathology with deep learning-assisted gland analysis.
_Cancer Res._ 82, 334–345 (2022). Article CAS PubMed MATH Google Scholar * Liu, J. T. C. et al. Harnessing non-destructive 3D pathology. _Nat. Biomed. Eng._ 5, 203–218 (2021). Article
PubMed PubMed Central MATH Google Scholar * iDISCO+ protocol. https://idisco.info/wp-content/uploads/2015/04/whole-mount-staining-bench-protocol-methanol-dec-2016.pdf. * Seo, J. et al.
PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements. _Nat. Commun._ 13, 1–17 (2022). Article MATH ADS Google
Scholar * Bai, B. et al. Deep learning-enabled virtual histological staining of biological samples. _Light Sci. Appl_ 12, 57 (2023). Article CAS PubMed PubMed Central MATH Google
Scholar * Suhling, K., French, P. M. W. & Phillips, D. Time-resolved fluorescence microscopy. _Photochem. Photobiol. Sci._ 4, 13–22 (2005). Article CAS PubMed Google Scholar *
Buchwalow, I., Samoilova, V., Boecker, W. & Tiemann, M. Multiple immunolabeling with antibodies from the same host species in combination with tyramide signal amplification. _Acta
Histochem._ 120, 405–411 (2018). Article CAS PubMed Google Scholar * Dahiya, V. & Chaudhuri, T. K. Chaperones GroEL/GroES accelerate the refolding of a multidomain protein through
modulating on-pathway intermediates. _J. Biol. Chem._ 289, 286–298 (2014). Article CAS PubMed MATH Google Scholar * Mai, H. et al. Scalable tissue labeling and clearing of intact human
organs. _Nat. Protoc._ 17, 2188–2215 (2022). Article CAS PubMed MATH Google Scholar * Voigt, F. F. et al. The mesoSPIM initiative: open-source light-sheet microscopes for imaging
cleared tissue. _Nat. Methods_ 16, 1105–1108 (2019). Article CAS PubMed PubMed Central MATH Google Scholar * Choi, H. M. T., Beck, V. A. & Pierce, N. A. Next-generation in situ
hybridization chain reaction: higher gain, lower cost, greater durability. _ACS Nano_ 8, 4284–4294 (2014). Article CAS PubMed PubMed Central Google Scholar * Tsuneoka, Y. & Funato,
H. Modified in situ hybridization chain reaction using short hairpin DNAs. _Front. Mol. Neurosci._ 13, 75 (2020). Article CAS PubMed PubMed Central Google Scholar * Hörl, D. et al.
BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. _Nat. Methods_ 16, 870–874 (2019). Article PubMed MATH Google Scholar * Arshadi, C., Günther,
U., Eddison, M., Harrington, K. I. S. & Ferreira, T. A. SNT: a unifying toolbox for quantification of neuronal anatomy. _Nat. Methods_ 18, 374–377 (2021). Article CAS PubMed Google
Scholar * Soille, P. _Morphological Image Analysis_ (Springer Berlin Heidelberg, 2004). * Bigun, J. Optimal orientation detection of linear symmetry. In _Proc. IEEE-First International
Conference on Computer Vision_. 433–438 (IEEE, London, 1987). * Khan, A. R. et al. 3D structure tensor analysis of light microscopy data for validating diffusion MRI. _Neuroimage_ 111,
192–203 (2015). Article PubMed MATH Google Scholar * Bigun, J., Bigun, T. & Nilsson, K. Recognition by symmetry derivatives and the generalized structure tensor. _IEEE Trans. Pattern
Anal. Mach. Intell._ 26, 1590–1605 (2004). Article PubMed MATH Google Scholar * Website. Connor Meehan, Jonathan Ebrahimian, Wayne Moore, and Stephen Meehan (2022). Uniform Manifold
Approximation and Projection (UMAP) (https://www.mathworks.com/matlabcentral/fileexchange/71902), MATLAB Central File Exchange. * Morel, P. Gramm: grammar of graphics plotting in Matlab. _J.
Open Source Softw._ 3, 568 (2018). Article MATH ADS Google Scholar Download references ACKNOWLEDGEMENTS We would like to express our deepest gratitude to the tissue donor and his family
for their generosity and wisdom in supporting scientific studies. We thank William Wu for access to confocal microscopy; Cathy Shuk Ling Chan for manual data annotation and analysis; Ka Wai
Chan for administrative support to the project. Figures 1and 6h were partly created with BioRender.com. The project was supported by the Midstream Research Program for Universities
(MRP/048/20, H.M.L.) of the Innovation and Technology Council of Hong Kong, a Direct Grant for Research 2022/23 (2022.072, H.M.L.) of the Chinese University of Hong Kong, and the Chinese
University of Hong Kong Research Committee Group Research Scheme (GRS) 2021–22 (granted to Y.K.W.). AUTHOR INFORMATION Author notes * These authors contributed equally: Chun Ngo Yau, Jacky
Tin Shing Hung. AUTHORS AND AFFILIATIONS * Department of Psychiatry, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China Chun Ngo Yau, Jacky Tin Shing Hung, Thomas
Chun Yip Wong, Bei Huang, Ben Tin Yan Wong, Nick King Ngai Chow, Lichun Zhang, Eldric Pui Lam Tsoi, Yun Kwok Wing & Hei Ming Lai * Li Ka Shing Institute of Health Sciences, Faculty of
Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China Chun Ngo Yau, Jacky Tin Shing Hung, Thomas Chun Yip Wong, Bei Huang, Ben Tin Yan Wong, Nick King Ngai Chow, Lichun Zhang,
Eldric Pui Lam Tsoi, Yun Kwok Wing & Hei Ming Lai * Sainsbury Wellcome Centre for Neural Circuits and Behaviour, University College London, London, UK Robert A. A. Campbell * Li Chiu
Kong Family Sleep Assessment Unit, Department of Psychiatry, The Chinese University of Hong Kong, Hong Kong SAR, China Bei Huang & Yun Kwok Wing * Department of Microbiology and
Immunology, Stanford University, Stanford, CA, USA Yuqi Tan * Department of Pathology, School of Clinical Medicine, The University of Hong Kong, Queen Mary Hospital, Hong Kong SAR, China
Joshua Jing Xi Li * Department of Chemical Pathology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China Hei Ming Lai Authors * Chun Ngo Yau View author
publications You can also search for this author inPubMed Google Scholar * Jacky Tin Shing Hung View author publications You can also search for this author inPubMed Google Scholar * Robert
A. A. Campbell View author publications You can also search for this author inPubMed Google Scholar * Thomas Chun Yip Wong View author publications You can also search for this author
inPubMed Google Scholar * Bei Huang View author publications You can also search for this author inPubMed Google Scholar * Ben Tin Yan Wong View author publications You can also search for
this author inPubMed Google Scholar * Nick King Ngai Chow View author publications You can also search for this author inPubMed Google Scholar * Lichun Zhang View author publications You can
also search for this author inPubMed Google Scholar * Eldric Pui Lam Tsoi View author publications You can also search for this author inPubMed Google Scholar * Yuqi Tan View author
publications You can also search for this author inPubMed Google Scholar * Joshua Jing Xi Li View author publications You can also search for this author inPubMed Google Scholar * Yun Kwok
Wing View author publications You can also search for this author inPubMed Google Scholar * Hei Ming Lai View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS Conceptualization: H.M.L. Methodology: C.N.Y., J.T.S.H., R.A.A.C., T.C.Y.W., B.T.Y.W., N.K.N.C., L.Z., E.P.L.T., H.M.L. Investigation: C.N.Y., J.T.S.H., R.A.A.C., T.C.Y.W.,
B.H., B.T.Y.W., N.K.N.C., L.Z., E.P.L.T., Y.T., J.J.X.L., Y.K.W., H.M.L. Visualization: C.N.Y., J.T.S.H., R.A.A.C., H.M.L. Funding acquisition: Y.K.W., H.M.L. Project administration: H.M.L.
Supervision: H.M.L. Writing: C.N.Y., H.M.L. CORRESPONDING AUTHOR Correspondence to Hei Ming Lai. ETHICS DECLARATIONS COMPETING INTERESTS C.U.H.K. filed a patent application in part based on
the invention described in this paper with H.M.L. and C.N.Y. as the inventors. The associated patent, owned by C.U.H.K., was exclusively licensed to Illumos Limited, of which H.M.L. is a
co-founder. The remaining authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks Ying Hu, who co-reviewed with Hirushi Gunasekara; Angela
Sirigu, who co-reviewed with Guillaume Lio; Irene Costantini; and Masaharu Yoshihara for their contribution to the peer review of this work. A peer review file is available. ADDITIONAL
INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY MOVIE 1 SUPPLEMENTARY MOVIE 2 SUPPLEMENTARY MOVIE 3 REPORTING SUMMARY TRANSPARENT PEER REVIEW FILE SOURCE DATA SOURCE
DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes
were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Yau, C.N., Hung, J.T.S., Campbell, R.A.A. _et al._ INSIHGT: an accessible multi-scale, multi-modal 3D spatial biology platform. _Nat Commun_ 15, 10888 (2024).
https://doi.org/10.1038/s41467-024-55248-0 Download citation * Received: 28 June 2024 * Accepted: 06 December 2024 * Published: 30 December 2024 * DOI:
https://doi.org/10.1038/s41467-024-55248-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Woman, 18, drowns in boating accidentAn 18-year-old Ojai woman drowned while boating with a group of friends on Lake Nacimiento in San Luis Obispo County, au...
Aarp asks the candidates: california 25th congressional districtMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...
Celtic and rangers on alert as uefa to make champions league rule changeEUROPEAN FOOTBALL’S GOVERNING BODY ARE REPORTEDLY SET TO MAKE A CHANGE TO THE FORMAT NEXT SEASON 08:36, 02 Jun 2025 Celt...
Locations | kalispell vet center | veterans affairsMAIN LOCATION 690 North Meridian Road Suite 101 Kalispell, MT 59901 * Mon. 8:00 a.m. to 4:30 p.m. * Tue. 8:00 a.m. to 4:...
Lauren silverman wows in boob-baring swimsuit but mum steals spotlightThe brunette beauty’s incredible figure was impossible to miss as she made a beeline for a speed boat as the famous fami...
Latests News
Insihgt: an accessible multi-scale, multi-modal 3d spatial biology platformABSTRACT Biological systems are complex, encompassing intertwined spatial, molecular and functional features. However, m...
Every day is veterans day | va poplar bluff health care | veterans affairsOn October 22, 2015, staff at the John J. Pershing VA Medical Center received a call from a Veteran’s wife. Her husband...
Special rajdhani express from new delhi to mumbai starts today: fares, routes and other detailsThe Delhi-Mumbai line is important for the railways as the route, once the reserve of business travellers, has been losi...
Arsenal boss mikel arteta names martin odegaard as future captainMikel Arteta believes that Martin Odegaard can be a future Arsenal captain. As Arsenal, inch by inch, have rebuilt their...
Edinburgh festivals: how they became the world’s biggest arts eventThe Edinburgh Festival is upon us again, a three-week spectacular that turns the Scottish capital into the biggest arts ...