Interleukin-17a stimulation induces alterations in microglial microrna expression profiles
Interleukin-17a stimulation induces alterations in microglial microrna expression profiles"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT BACKGROUND Increased maternal interleukin (IL)-17A and activated microglia are pivotal factors contributing to the pathological phenotypes of maternal immune activation (MIA),
developing neurodevelopmental disorders in offspring. This study aimed to determine whether IL-17A affects the microglial microRNA (miRNA) profiles. METHODS The miRNA expression profiles of
primary cultured microglia stimulated with recombinant IL-17A were examined comprehensively using miRNA sequencing and validated through qRT-PCR. The expressions of miRNAs target genes
identified using bioinformatics, were investigated in microglia transfected with mimic miRNA. The target gene’s expression was also examined in the fetal brains of the MIA mouse model
induced by maternal lipopolysaccharide (LPS) administration. RESULTS Primary cultured microglia expressed the IL-17A receptor and increased proinflammatory cytokines and _nitric oxide
synthase 2_ upon treatment with IL-17A. Among the three miRNAs with |log2FC | >1, only mmu-miR-206-3p expression was significantly up-regulated by IL-17A. Transfection with the
mmu-miR-206-3p mimic resulted in a significant decrease in the expression of _Hdac4_ and _Igf1_, target genes of mmu-miR-206-3p. _Hdac4_ expression also significantly decreased in the
LPS-induced MIA model. CONCLUSIONS IL-17A affected microglial miRNA profiles with upregulated mmu-miR-206-3p. These findings suggest that targeting the IL-17A/mmu-miR-206-3p pathway may be a
new strategy for predicting MIA-related neurodevelopmental deficits and providing preventive interventions. IMPACT * Despite the growing evidence of interleukin (IL)-17A and microglia in
the pathology of maternal immune activation (MIA), the downstream of IL-17A in microglia is not fully known. * IL-17A altered microRNA profiles and upregulated the mmu-miR-206-3p expression
in microglia. The mmu-miR-206-3p reduced autism spectrum disorder (ASD) related gene expressions, _Hdac4_ and _Igf1_. * The _Hdac4_ expression was also reduced in the brain of MIA offspring.
* The hsa-miR-206 sequence is consistent with that of mmu-miR-206-3p. * This study may provide clues to pathological mechanisms leading to predictions and interventions for ASD children
born to mothers with IL-17A-related disorders. You have full access to this article via your institution. Download PDF SIMILAR CONTENT BEING VIEWED BY OTHERS MATERNAL IMMUNE ACTIVATION
ALTERS FETAL AND NEONATAL MICROGLIA PHENOTYPE AND DISRUPTS NEUROGENESIS IN MICE Article 13 August 2022 MATERNAL IMMUNE ACTIVATION INDUCES SUSTAINED CHANGES IN FETAL MICROGLIA MOTILITY
Article Open access 07 December 2020 MODULATING MICROGLIA ACTIVATION PREVENTS MATERNAL IMMUNE ACTIVATION INDUCED SCHIZOPHRENIA-RELEVANT BEHAVIOR PHENOTYPES VIA ARGINASE 1 IN THE DENTATE
GYRUS Article 29 June 2020 INTRODUCTION Emerging evidence suggests that maternal immune activation (MIA) plays a crucial role in causing neurodevelopmental disorders, including autism
spectrum disorder (ASD)1,2. ASD is known to be associated with both environmental and genetic factors3, and MIA is an important environmental factor _in utero_. Currently, interleukin (IL)
-17A and microglial activation have been reported to play pivotal roles in MIA pathology1,4: (i) IL-6, induced by infection or immunological stimulation, stimulates maternal intestinal T
helper 17 (Th17) cells, leading to the secretion of IL-17A; (ii) IL-17A crosses the placenta, resulting in inflammatory alterations in the fetal brain _in utero_, including microglial
activation; and (iii) offspring show the behavioral and neuronal deficits observed in ASD and architectural changes in the brain consistent with the findings observed in grown-up state.
Blocking IL-17A with antibodies has also been shown to attenuate ASD-like behavior in an poly(I:C)-induced MIA model5. Microglial activation is a critical process in MIA pathogenesis. We
have previously reported that microglial activation in the fetal brain is associated with ASD-like behavior using a lipopolysaccharide (LPS)-induced MIA model6,7. Fetal microglia derived
from the yolk sac support and regulate neurogenesis by releasing neurotrophic factors, including insulin growth factor 1 (IGF1), promoting neural precursor cell proliferation, myelination,
and interneuron wiring8. However, signals of MIA, mainly IL-17A, cross the immature blood-brain barrier in the fetal brain, and cause microglial activation or abnormal microglial behavior9.
Microglia have a functional IL-17A receptor (IL-17RA) and respond to IL-17A stimulation9,10. Microglia decrease the expression of molecules involved in supporting interneuron development in
an IL-17A dependent manner11. Thus, the downstream of IL-17A in microglia is suggested to be a potential target for MIA8. However, the effects of IL-17A on microglial phenotype remain
incompletely understood. MicroRNAs (miRNAs) are small non-coding RNAs that are involved in the regulation of post-transcriptional gene expression. They play a role in regulating the
magnitude of inflammatory responses in the immune cells12 and expression of miRNAs has also been implicated in microglial function and neurodevelopmental disorders following prenatal
exposure13. An increased expression of miR-155 has been reported in the postmortem brain tissues of children with ASD14, and miRNA profiles have been shown to be altered in the brains of MIA
mouse model15. Based on these observations, in this study, we focused on miRNAs in microglia that may be related to the development of neurodevelopmental disorders by altering microglial
phenotypes. We aimed to identify the miRNAs showing altered expression in microglia following IL-17A stimulation. MATERIALS AND METHODS REAGENTS Lipopolysaccharide (LPS, serotype O55:B5) was
purchased from Sigma-Aldrich (St. Louis, MO). Recombinant murine IL-17A was purchased from PeproTech (Cat No. 210-17: Rocky Hill, NJ). ANIMALS The experimental protocols in this study were
approved by the Animal Experiment Committee of Nagoya University (approval number: M220211-001) and were carried out in accordance with the Regulations on Animal Experiments at Nagoya
University. Pregnant Slc:ICR mice were purchased from Japan SLC (Shizuoka, Japan). All mice were provided with free access to food (CE-2; CLEA Japan, Inc., Tokyo, Japan) and water and were
maintained on a 12 h light/12 h dark lighting (lights on at 9:00 AM). Pregnant mice (_n_ = 8) were randomly divided into two groups. The mice in the LPS-induced MIA group received an
intraperitoneal injection (i.p.) of 50 μg LPS dissolved in phosphate-buffered saline (PBS) on embryonic day 17 (E17), while the control group received an equal volume of sterile PBS on the
same day (_n_ = 4 in each group). The maternal blood samples (_n_ = 4 in each group) and fetal brains (_n_ = 3 in each group) were collected from the two groups 8 h later of PBS or LPS i.p.
under deep anesthesia. IL-17A MEASUREMENT IL-17A levels in the maternal serum were determined using an ELISA kit (Mouse IL-17A ELISA Kit # ab199081; Abcam, Cambridge, UK). Absorbance at 450
nm was measured using a multiple plate reader according to the manufacturer’s protocol (Thermo ScientificTM MultiskanTM FC; Thermo Fisher Scientific Inc., Waltham, MA). PREPARATION OF
PRIMARY MOUSE MICROGLIA Primary microglia were isolated from primary mixed glial cell cultures prepared from newborn ICR mice on day 14 using the ‘shaking off’ method as described
previously6,16. The purity of the cultures (>99%) was determined using anti-CD11b immunostaining (BD Biosciences, Franklin Lakes, NJ). Cultures were maintained in Dulbecco’s modified
Eagle’s minimum essential medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Equitech-Bio, Kerrville, TX), 5 mg/mL bovine insulin (Sigma-Aldrich), and 0.2% glucose. Primary
microglia were plated at 2 × 105 cells/well in 1 mL of culture medium in 24-well plates and were incubated for 24 h at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air. Either
0 or 10 ng/mL of IL-17A was added to each well and incubated for 24 h at 37 °C. The concentration of IL-17A was determined according to a previous report10. MIRNA PROFILING miRNA profiling
was performed as previously described17. Briefly, total miRNA was isolated from microglial cultures using the miRNeasy Mini Kit (Qiagen Inc., Hilden, Germany) following the manufacturer’s
instructions. Small RNA libraries were prepared, PCR products were purified, DNA fragments were recovered, and cDNA concentration was measured as previously reported17. Single-end reads were
obtained using Illumina MiSeq (Illumina, San Diego, CA). Data analysis was performed using the CLC Genomics Workbench version 9.5.3 program (Qiagen Inc.), as previously reported17. After
the adaptor sequences were trimmed, sequences from 15 to 35 bp were counted. The data were mapped to the miRBase 21 database, allowing for a maximum of up to two mismatches18. Normalization
was performed using reads per million mapped reads, followed by exclusion of low-expression (<100 reads in control samples) miRNAs (Fig. 2a). miRNAs with |log2FC | > 0.6 were selected,
and their target genes were predicted using TargetScanMouse (release 8.0, September 2021, https://www.targetscan.org/mmu_80/), and duplicate genes were removed. The selected genes were used
for Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on Metascape (http://metascape.org), as previously reported19,20. QUANTITATIVE REVERSE
TRANSCRIPTION-POLYMERASE CHAIN REACTION (QRT-PCR) OF MIRNA qRT-PCR was conducted using primary cultured microglia stimulated with or without 10 ng/mL IL-17A. Total miRNA was isolated from
primary cultured microglia (n = 3–4 in each group) using the miRNeasy Mini Kit (Qiagen Inc.) following the manufacturer’s instructions. RNA concentration and RNA integrity number were
measured using Agilet 2100 Bioanalyzer with Agilet 6000 RNA pico kit (Agilent Technologies, Santa Clara, CA). Briefly, cDNA was synthesized using the TaqMan Advanced miRNA cDNA Synthesis Kit
(Thermo Fisher Scientific Inc.) according to the manufacturer’s instructions. TaqMan® Fast Advanced Master Mix (Thermo Fisher Scientific Inc.) and TaqMan™ Advanced miRNA Assay (Assay ID
481645_mir for mmu-miR-206-3p, Assay ID 482687_mir for miR-151-5p, Assay ID 481300_mir for mmu-miR-29b-3p, Assay ID 481173_mir for mmu-miR-455-5p, Assay ID 481709_mir for mmu-miR-434-5p,
Assay ID 482619_mir for mmu-miR-28a-5p, Assay ID 478254_mir for mmu-miR-107-3p, Assay ID 481485_mir for mmu-miR-181a-5p, Assay ID 481004_mir for mmu-miR-22-3p, Assay ID 478587_mir for
hsa-miR-29a-3p, Assay ID 478412_mir for mmu-miR-106b-5p, and Assay ID 482926_mir for mmu-miR-381-3p) were used. Then, qRT-PCR was performed using a Thermal Cycler Dice (Takara Bio Inc.,
Tokyo, Japan), and the PCR conditions included denaturation at 95 °C for 10 min, followed by 45 amplification cycles of 95 °C for 15 s and 60 °C for 1 min. The amplified product was
monitored by measuring the increase in the FAM (fluorescein) fluorescence intensity. TRANSFECTION OF MIRNA MIMICS Primary microglia were plated in 24-well plates (2 × 105 cells/well). The
ectopic mmu-miR‐206-3p expression was introduced into the cells by transfection with double-strand-mmu-miR-206-3p (5ʹ-UGGAAUGUAAGGAAGUGUGUGG-3ʹ 50 nM; Bioneer S-1017-1) using RNAiMAX (Thermo
Fisher Scientific Inc.), according to the manufacturer’s instructions. Negative control mimic (Accutarget miRNA mimic negative control; SMC-2002, Bioneer Inc. Oakland, CA) was used as a
negative control (50 nM). Following incubation for 12 h, the half of culture medium was changed, and cells were incubated for an additional 12 h. RT-PCR AND QRT-PCR OF MRNA Total RNA was
isolated from primary cultured microglia (_n_ = 3–8) and the fetal brains (_n_ = 3) using the RNeasy Mini Kit (Qiagen Inc.), following the manufacturer’s instructions. The total RNA
concentration was measured using a Nanodrop (Thermo Fisher Scientific Inc.). The RT reaction with 300 ng of total RNA was performed using a first-strand synthesis kit (ReverTra Ace; Toyobo
Co., Ltd., Osaka, Japan). RT-PCR was performed on the cDNA of primary cultured microglia using a Veriti Thermal Cycler (Thermo Fisher Scientific Inc.) with Blend taq (Toyobo Co., Ltd.), as
previously reported21. PCR conditions were 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 sec, annealing at 60 °C for 30 s, and extension at 72 °C for 1 min. The
amplification products were electrophoresed on 15% polyacrylamide gels. qRT-PCR was performed in 96-well PCR plates using Thermal Cycler Dice (Takara Bio Inc.) and KOD SYBR qPCR Mix (Toyobo
Co., Ltd.) reagents. The primers used for the RT- and qRT-PCR are listed in Table 1. _Actb_ was used as an endogenous reference gene. Relative expression in the IL-17A group was determined
by setting the values in the control group to 1.0. STATISTICAL ANALYSIS The data are presented as the mean ± standard error of the mean (SEM) and were analyzed using the Student’s _t_ test.
Statistical analysis was performed using Prism version 8.4.3 Windows (GraphPad Software Inc., San Diego, CA). Statistical significance was set at _p_ < 0.05. RESULTS IL-17A-INDUCED
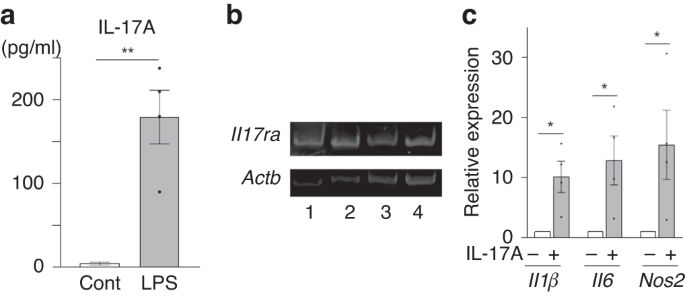
INFLAMMATORY RESPONSE IN PRIMARY MICROGLIA CELLS The serum levels of IL-17A in the LPS-induced MIA group were significantly higher than those in the control group (mean ± SEM, 179.5 ± 32.1
vs. 3.9 ± 1.8 pg/mL, _p_ < 0.01, Fig. 1a). Primary cultured microglia revealed the expression of IL-17A receptor (_Il17ra_) (Fig. 1b). The mRNA expression of _IL1β, IL6_, and nitric oxide
synthase 2 (_Nos2_), as activated microglial molecules, was increased significantly with IL-17A treatment (_p_ < 0.05, Fig. 1c). IL-17A INDUCED ALTERATIONS IN MIRNA PROFILES After
normalization, 166 miRNAs were detected in more than 100 reads (Fig. 2a). A total of 3057 genes were extracted as target genes of 12 miRNAs (|log2FC | > 0.6), and 2805 annotated genes
were identified among them through functional annotation analysis (Fig. 2a). The top pathway was identified as the pathway for the regulation of neuronal differentiation, and other relevant
pathways, including developmental growth, head development, histone modification, in utero embryonic development, dendrite development, axon guidance, and behavior, were listed in the top 20
dysregulated pathways (Fig. 2b). Twelve miRNAs with |log2FC | > 0.6 were detected (Fig. 3a). Of these, mmu-miR-206-3p was upregulated (log2FC > 1) and mmu-miR-29b-3p and
mmu-miR-151-5p were downregulated (log2FC < −1) in the IL-17A group compared to the control group (Fig. 3a). The validation of three miRNAs (mmu-miR-206-3p, mmu-miR-29b-3p, and
mmu-miR-151-5p) revealed that mmu-miR-206-3p was significantly upregulated (_p_ < 0.05), while mmu-miR-29b-3p and mmu-miR-151-5p showed no significant difference (Fig. 3b). Differentially
expressed miRNAs with 1.0 ≥ |log2FC | > 0.6 by IL-17A were also validated (Fig. S1). Only mmu-miR-381-3p was significantly downregulated (_p_ < 0.05), but the expression levels were
low in both groups. The other eight miRNAs showed no significant difference. IDENTIFICATION OF THE TARGET GENES OF MMU-MIR-206-3P A total of 729 genes were identified as putative target
genes of mmu-miR-206-3p. Among these genes, brain-derived neurotrophic factor (_Bdnf_), histone deacetylase 4 (_Hdac4_), and _Igf1_ were focused on because they are well-known for their
functions in microglia and epigenetics. They contain the binding sites for mmu-miR-206-3p in their 3′-untranslated regions (Fig. 4a). The hsa-miR-206 showed the sequence that was identical
to that of mmu-miR-206-3p, and similar binding sites were identified in _Bdnf_, _Hdac4_, and _Igf1_ (Fig. 4b). The mmu-miR-206-3p mimic was successfully transfected into the microglia (Fig.
4c). Transfection of the mmu-miR-206-3p mimic significantly decreased _Hdac4_ and _Igf1_ expression (_p_ < 0.01 and 0.05, respectively, Fig. 4d), but had no significant effect on _Bdnf_
transcript levels (Fig. 4d). IL-17A INDUCED REDUCTION OF _IGF1_ IN MICROGLIA The effect of IL-17A was examined on target genes of mmu-miR-206-3p, _Bdnf_, _Hdac4_, and _Igf1_. IL-17A
significantly decreased the _Igf1_ expression (_p_ < 0.05, Fig. 5a). _Hdac4_ were also decreased by IL-17A, although they were not significant (_p_ = 0.39, Fig. 5a). In addition, _Itgax_
expression, a microglial marker, also known as _Cd11c_, was reduced by IL-17A treatment (_p_ < 0.05, Fig. 5b). REDUCED _HDAC4_ LEVELS IN THE FETAL BRAINS OF THE LPS-INDUCED MIA MOUSE
MODEL In the LPS group, _Hdac4_ expression in the fetal brains was significantly reduced compared to that in the control group (_p_ < 0.05, Fig. 5c). However, _Igf1_ expression in the LPS
group was not significantly different from that in the control group (_p_ = 0.61, Fig. 5c). DISCUSSION To the best of our knowledge, the present study is the first to demonstrate that
IL-17A alters miRNA profiles in microglia with significantly upregulated mmu-miR-206-3p. We found that the expression levels of _Hdac4_ and _Igf1_ were negatively regulated by
mmu-miR-206-3p, and _Igf1_ expression was reduced by IL-17A in microglia. Moreover, _Hdac4_ expression was reduced in the brains of the offspring, and maternal IL-17A was increased in the
LPS-induced MIA model. However, IL-17A slightly affected _Hdac4_ expression in microglia. These findings suggest that IL-17A may be involved in the pathogenesis of the LPS-induced MIA model
by inducing mmu-miR-206-3p in the microglia. Notably, the hsa-miR-206 sequence was consistent with that of mmu-miR-206-3p, and the results in this study would be similar for human microglia.
The present study is the first to demonstrate that mmu-miR-206-3p was induced by IL-17A, a critical molecule in MIA, although a correlation between IL-17A and hsa-miR-206 was previously
shown22. Previous studies have reported that mmu-miR-206-3p and hsa-miR-206 have pathophysiological roles in neuropsychiatric disorders, including anxiety or depression23,24,25, and
Alzheimer’s disease26,27,28. Intranasal administration of AgomiR-206-3p and AntagomiR-206-3p (AM206) has shown induced depressive-like behaviors and antidepressant-like effects in mouse
models, respectively23. MIA can also cause anxiety disorders or depression29. These findings suggested that mmu-miR-206-3p may also be a critical factor in neuropsychiatric disorders
associated with MIA. The mmu-miR-206-3p and hsa-miR-206 have also been known to regulate the _IGF1_ expression in various other cells, including trophoblasts and muscle cells30,31,32.
Moreover, hsa/mml/mmu-miR-206 is known to be involved in the regulation of _Hdac4_ as a target gene in muscle cells33,34,35,36 and hsa-miR-206 in neurons37. Furthermore, the
hsa-miR-133b/miR-206 variants in the cluster (rs16992131) are found in the lymphocytes of patients with ASD compared to those from healthy controls38. Schizophrenia, which shares
pathophysiology with ASD, is also associated with hsa-miR-20639,40,41. The rno-miR-206-3p is upregulated in the hippocampus of attention-deficit/hyperactivity disorder rats42. Those findings
highlight their importance in neurodevelopmental disorders. Recently, IGF1 has been suggested to play a role in neuroprotection and neurodevelopment, and proposed to have a potential in
aiding ASD treatment43,44,45,46. Moreover, CD11c-positive microglia have been shown to regulate myelination by producing IGF147. The results obtained in the present study showing a decrease
in the _Itgax_ (_Cd11c_) and _Igf1_ expression in microglia following IL-17A treatment, were consistent with those findings. This study supports the therapeutic potential of IGF1 for ASD
caused by maternal IL-17A-related inflammation. A previous study reported that LPS decreases IGF1 in the hippocampus but not in the frontal cortex after 3 h of the injection48. However, no
decrease in _Igf1_ was observed in the whole brain of our LPS-induced MIA model, which may be due to the site and timing of sampling. HDAC4 is a member of the HDAC family that epigenetically
regulates gene transcription by interacting with HDAC3 or other transcriptional factors. _Hdac4_ is highly expressed in the brain49 and plays a pivotal role in neurodevelopment and
cognitive function50. The present study demonstrated that _Hdac4_ levels were reduced in the LPS-induced MIA model. This model indicates that short-term memory and social interactions are
impaired, as we previously reported7. Thus, reduced _Hdac4_ levels may be associated with those neurodevelopment deficits. However, IL-17A slightly reduced _Hdac4_ expression in microglia,
although not significant. Those findings suggest that mmu-miR-206-3p could reduce _Hdac_4 in microglia, but the reduction of _Hdac_4 in the LPS-induced MIA model might be partly associated
with the IL-17A/mmu-miR-206-3p pathway. The strength of this study is that it is the first to show that IL-17A alters the miRNA profile and increases mmu-miR-206-3p in primary cultured
microglia. The results from primary cultured microglia are considered valuable since reactions of microglia cell lines are often inconsistent with those of primary cultured microglia51, 52.
The present results suggest that maternal IL-17A increases mmu-miR-206-3p in the microglia of their offspring, which may be related to the pathogenesis of the LPS-induced MIA model. However,
whether several pathways, including _Igf1_, are involved in ASD-like behavior should be further investigated. Additionally, hsa-miR-206 may play a critical role in developing ASD in
children born to a mother with IL-17A-related pathology since it showed an identical sequence to mmu-miR-206-3p. Thus, new management strategies should also be established to protect fetal
microglia from maternal IL-17A. Indeed, in Alzheimer’s disease, hsa-miR-206 has been considered useful as a possible predictor of progression and early diagnosis26,27. Furthermore, AM206 has
been reported to be a potential therapeutic agent for Alzheimer’s disease28 and stress-exacerbated aggressive behavior53. This study has several limitations. First, we used mouse-derived
microglia, and the miRNA profiles may not be applicable to human microglia. However, the sequence of hsa-miR-206 was consistent with that of mmu-miR-206-3p and had similar HDAC4 and IGF1
binding sites. Therefore, to validate our results, further investigations using human samples are required. Second, we did not evaluate whether mmu-miR-206-3p is involved in microglial
phenotypes, including synapse pruning. A previous study reported that the mmu-miR-206 can enhance amyloidogenesis by targeting IGF1 in microglia, leading to Alzheimer’s disease54, but
further investigation is needed to identify the role of mmu-miR-206 in microglial function. Third, further studies are required to identify the pathways that IL-17A reduces the _Igf1_ via
mmu-miR-206-3p. Despite these limitations, our study suggests that mmu-miR-206-3p might be associated with the pathogenesis of MIA, although further studies with MIA mouse models treated
with AM206 are needed. CONCLUSIONS In conclusion, we have demonstrated that higher maternal IL-17A levels can lead to changes in microglial miRNAs with upregulated mmu-miR-206-3p. IL-17A can
modify gene expressions, including _Igf1_, in microglia via the mmu-miR-206-3p, although further investigation is needed to confirm that. These findings suggest that targeting the
mmu-miR-206-3p pathway may be a useful predictive and preventive intervention strategy for neurodevelopmental disorders associated with MIA. DATA AVAILABILITY The datasets generated and
analysed during the current study are available in the DDBJ Sequence Read Archive (DRA015999). All other relevant data are available within the article file from the authors upon reasonable
request. REFERENCES * Estes, M. L. & McAllister, A. K. Maternal immune activation: Implications for neuropsychiatric disorders. _Science_ 353, 772–777 (2016). Article CAS PubMed
PubMed Central Google Scholar * Knuesel, I. et al. Maternal immune activation and abnormal brain development across Cns disorders. _Nat. Rev. Neurol._ 10, 643–660 (2014). Article CAS
PubMed Google Scholar * Taylor, M. J. et al. Etiology of autism spectrum disorders and autistic traits over time. _JAMA Psychiatry_ 77, 936–943 (2020). Article PubMed Google Scholar *
Wong, H. & Hoeffer, C. Maternal Il-17a in autism. _Exp. Neurol._ 299, 228–240 (2018). Article CAS PubMed Google Scholar * Choi, G. B. et al. The maternal interleukin-17a pathway in
mice promotes autism-like phenotypes in offspring. _Science_ 351, 933–939 (2016). Article CAS PubMed PubMed Central Google Scholar * Imai, K. et al. Neuroprotective potential of
molecular hydrogen against perinatal brain injury via suppression of activated microglia. _Free Radic. Biol. Med._ 91, 154–163 (2016). Article CAS PubMed Google Scholar * Imai, K. et al.
Administration of molecular hydrogen during pregnancy improves behavioral abnormalities of offspring in a maternal immune activation model. _Sci. Rep._ 8, 9221 (2018). Article PubMed
PubMed Central Google Scholar * Otero, A. M. & Antonson, A. M. At the crux of maternal immune activation: Viruses, microglia, microbes, and Il-17a. _Immunol. Rev._ 311, 205–223 (2022).
Article CAS PubMed PubMed Central Google Scholar * Das Sarma, J. et al. Functional interleukin-17 receptor a is expressed in central nervous system glia and upregulated in experimental
autoimmune encephalomyelitis. _J. Neuroinflammation_ 6, 14 (2009). Article PubMed PubMed Central Google Scholar * Kawanokuchi, J. et al. Production and functions of Il-17 in Microglia.
_J. Neuroimmunol._ 194, 54–61 (2008). Article CAS PubMed Google Scholar * Yu, D. et al. Microglial Gpr56 is the molecular target of maternal immune activation-induced
parvalbumin-positive interneuron deficits. _Sci. Adv._ 8, eabm2545 (2022). Article CAS PubMed PubMed Central Google Scholar * O’Connell, R. M., Rao, D. S. & Baltimore, D. Microrna
regulation of inflammatory responses. _Annu Rev. Immunol._ 30, 295–312 (2012). Article PubMed Google Scholar * Komada, M. & Nishimura, Y. Epigenetics and neuroinflammation associated
with neurodevelopmental disorders: A microglial perspective. _Front Cell Dev. Biol._ 10, 852752 (2022). Article PubMed PubMed Central Google Scholar * Almehmadi, K. A., Tsilioni, I.
& Theoharides, T. C. Increased expression of Mir-155p5 in amygdala of children with autism spectrum disorder. _Autism Res_ 13, 18–23 (2020). Article PubMed Google Scholar * Sunwoo, J.
S. et al. Maternal immune activation alters brain microrna expression in mouse offspring. _Ann. Clin. Transl. Neurol._ 5, 1264–1276 (2018). Article CAS PubMed PubMed Central Google
Scholar * Suzumura, A., Sawada, M. & Takayanagi, T. Production of interleukin-12 and expression of its receptors by murine microglia. _Brain Res_ 787, 139–142 (1998). Article CAS
PubMed Google Scholar * Yoshida, K. et al. Unique mirna profiling of squamous cell carcinoma arising from ovarian mature teratoma: Comprehensive mirna sequence analysis of its molecular
background. _Carcinogenesis_ 40, 1435–1444 (2019). CAS PubMed Google Scholar * Griffiths-Jones, S., Saini, H. K., van Dongen, S. & Enright, A. J. Mirbase: Tools for microrna genomics.
_Nucleic Acids Res_ 36, D154–D158 (2008). Article CAS PubMed Google Scholar * Kang, J. et al. Identification of key micrornas in diabetes mellitus erectile dysfunction rats with stem
cell therapy by bioinformatic analysis of deep sequencing data. _World J. Mens. Health_ 40, 663–677 (2022). Article PubMed PubMed Central Google Scholar * Zhang, L., Lu, D., Liu, M.,
Zhang, M. & Peng, Q. Identification and interaction analysis of key mirnas in medullary thyroid carcinoma by bioinformatics analysis. _Mol. Med Rep._ 20, 2316–2324 (2019). CAS PubMed
PubMed Central Google Scholar * Mizuno, M. et al. The role of E2f8 in the human placenta. _Mol. Med. Rep._ 19, 293–301 (2019). CAS PubMed Google Scholar * ElAtta, A. A., Ali, Y.,
Bassyouni, I. & Talaat, R. Correlation of myomir-206 and proinflammatory cytokines (Il-16 and Il-17) in patients with rheumatoid arthritis. _Reumatologia_ 57, 72–77 (2019). Article
PubMed PubMed Central Google Scholar * Guan, W. et al. Hippocampal Mir-206-3p participates in the pathogenesis of depression via regulating the expression of Bdnf. _Pharm. Res._ 174,
105932 (2021). Article CAS Google Scholar * Li, Q., Zhang, J., Gao, Z., Zhang, Y. & Gu, J. Gut microbiota-induced microrna-206-3p increases anxiety-like behaviors by inhibiting
expression of Cited2 and Stk39. _Micro. Pathog._ 176, 106008 (2023). Article CAS Google Scholar * Miao, Z. et al. Anxiety-related behaviours associated with microrna-206-3p and Bdnf
expression in pregnant female mice following psychological social stress. _Mol. Neurobiol._ 55, 1097–1111 (2018). Article CAS PubMed Google Scholar * Xie, B. et al. Increased serum
Mir-206 level predicts conversion from amnestic mild cognitive impairment to Alzheimer’s disease: A 5-year follow-up study. _J. Alzheimers Dis._ 55, 509–520 (2017). Article CAS PubMed
Google Scholar * Moon, J. et al. Early diagnosis of Alzheimer’s disease from elevated olfactory mucosal Mir-206 level. _Sci. Rep._ 6, 20364 (2016). Article CAS PubMed PubMed Central
Google Scholar * Lee, S. T. et al. Mir-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. _Ann. Neurol._ 72, 269–277 (2012). Article CAS PubMed Google Scholar *
Quagliato, L. A., de Matos, U. & Nardi, A. E. Maternal immune activation generates anxiety in offspring: A translational meta-analysis. _Transl. Psychiatry_ 11, 245 (2021). Article
PubMed PubMed Central Google Scholar * Sullivan, B. P. et al. Skeletal muscle Igf-1 is lower at rest and after resistance exercise in humans with obesity. _Eur. J. Appl. Physiol._ 120,
2835–2846 (2020). Article CAS PubMed Google Scholar * Wu, H. Y., Wang, X. H., Liu, K. & Zhang, J. L. Lncrna Malat1 regulates trophoblast cells migration and invasion Via
Mir-206/Igf-1 axis. _Cell Cycle_ 19, 39–52 (2020). Article CAS PubMed Google Scholar * Yu, Q., Zhao, B., He, Q., Zhang, Y. & Peng, X. B. Microrna-206 is required for osteoarthritis
development through its effect on apoptosis and autophagy of articular chondrocytes via modulating the phosphoinositide 3-kinase/protein kinase B-Mtor pathway by targeting insulin-like
growth factor-1. _J. Cell Biochem._ 120, 5287–5303 (2019). Article CAS PubMed Google Scholar * Simon, L. et al. Decreased myoblast differentiation in chronic binge alcohol-administered
simian immunodeficiency virus-infected male macaques: Role of decreased Mir-206. _Am. J. Physiol. Regul. Integr. Comp. Physiol._ 313, R240–r250 (2017). Article CAS PubMed PubMed Central
Google Scholar * Wen, J., He, T., Qi, F. & Chen, H. Mir-206-3p alleviates chronic constriction injury-induced neuropathic pain through targeting Hdac4. _Exp. Anim._ 68, 213–220 (2019).
Article CAS PubMed Google Scholar * Di Pietro, L. et al. Potential therapeutic targets for Als: Mir206, Mir208b and Mir499 are modulated during disease progression in the skeletal muscle
of patients. _Sci. Rep._ 7, 9538 (2017). Article PubMed PubMed Central Google Scholar * Pan, Y. et al. Activation of Ampk inhibits Tgf-Β1-induced airway smooth muscle cells
proliferation and its potential mechanisms. _Sci. Rep._ 8, 3624 (2018). Article PubMed PubMed Central Google Scholar * Guida, N. et al. The Mir206-Jund circuit mediates the neurotoxic
effect of methylmercury in cortical neurons. _Toxicol. Sci._ 163, 569–578 (2018). Article CAS PubMed Google Scholar * Toma, C. et al. Common and rare variants of microrna genes in autism
spectrum disorders. _World J. Biol. Psychiatry_ 16, 376–386 (2015). Article PubMed Google Scholar * Du, Y. et al. Genome-wide, integrative analysis implicates exosome-derived microrna
dysregulation in Schizophrenia. _Schizophr. Bull._ 45, 1257–1266 (2019). Article PubMed PubMed Central Google Scholar * Hansen, T. et al. Brain expressed micrornas implicated in
Schizophrenia etiology. _PLoS One_ 2, e873 (2007). Article PubMed PubMed Central Google Scholar * Hauberg, M. E. et al. Schizophrenia risk variants affecting microrna function and
site-specific regulation of Nt5c2 by Mir-206. _Eur. Neuropsychopharmacol._ 26, 1522–1526 (2016). Article CAS PubMed Google Scholar * Tian, T. et al. Mirna profiling in the hippocampus of
attention-deficit/hyperactivity disorder rats. _J. Cell Biochem_ 120, 3621–3629 (2019). Article CAS PubMed Google Scholar * Kolevzon, A. et al. Clinical trial of insulin-like growth
factor-1 in phelan-mcdermid syndrome. _Mol. Autism_ 13, 17 (2022). Article CAS PubMed PubMed Central Google Scholar * Linker, S. B., Mendes, A. P. D. & Marchetto, M. C. Igf-1
treatment causes unique transcriptional response in neurons from individuals with idiopathic autism. _Mol. Autism_ 11, 55 (2020). Article CAS PubMed PubMed Central Google Scholar *
Arjunan, A., Sah, D. K., Woo, M. & Song, J. Identification of the molecular mechanism of insulin-like growth factor-1 (Igf-1): A promising therapeutic target for neurodegenerative
diseases associated with metabolic syndrome. _Cell Biosci._ 13, 16 (2023). Article CAS PubMed PubMed Central Google Scholar * Costales, J. & Kolevzon, A. The therapeutic potential
of insulin-like growth factor-1 in central nervous system disorders. _Neurosci. Biobehav. Rev._ 63, 207–222 (2016). Article CAS PubMed PubMed Central Google Scholar * Wlodarczyk, A. et
al. A novel microglial subset plays a key role in myelinogenesis in developing brain. _EMBO J._ 36, 3292–3308 (2017). Article CAS PubMed PubMed Central Google Scholar * Szczesny, E. et
al. The impact of prenatal stress on insulin-like growth factor-1 and pro-inflammatory cytokine expression in the brains of adult male rats: The possible role of suppressors of cytokine
signaling proteins. _J. Neuroimmunol._ 276, 37–46 (2014). Article CAS PubMed Google Scholar * Grozinger, C. M., Hassig, C. A. & Schreiber, S. L. Three proteins define a class of
human histone deacetylases related to yeast Hda1p. _Proc. Natl. Acad. Sci. USA_ 96, 4868–4873 (1999). Article CAS PubMed PubMed Central Google Scholar * Wu, Y. et al. Aberrant
expression of histone deacetylases 4 in cognitive disorders: Molecular mechanisms and a potential target. _Front Mol. Neurosci._ 9, 114 (2016). Article PubMed PubMed Central Google
Scholar * Horvath, R. J., Nutile-McMenemy, N., Alkaitis, M. S. & Deleo, J. A. Differential migration, Lps-induced cytokine, chemokine, and no expression in immortalized Bv-2 and hapi
cell lines and primary microglial cultures. _J. Neurochem._ 107, 557–569 (2008). Article CAS PubMed PubMed Central Google Scholar * Henn, A. et al. The suitability of Bv2 cells as
alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. _Altex_ 26, 83–94 (2009). Article PubMed Google Scholar * Chang, C. H.,
Kuek, E. J. W., Su, C. L. & Gean, P. W. Microrna-206 regulates stress-provoked aggressive behaviors in post-weaning social isolation mice. _Mol. Ther. Nucleic Acids_ 20, 812–822 (2020).
Article CAS PubMed PubMed Central Google Scholar * Xing, H., Guo, S., Zhang, Y., Zheng, Z. & Wang, H. Upregulation of microrna-206 enhances lipopolysaccharide-induced inflammation
and release of amyloid-Β by targeting insulin-like growth factor 1 in microglia. _Mol. Med. Rep._ 14, 1357–1364 (2016). Article CAS PubMed Google Scholar Download references
ACKNOWLEDGEMENTS We express our gratitude to the members of the Department of Obstetrics and Gynecology at the Nagoya University Graduate School of Medicine. We received technical support
from the Division of Medical Research Engineering of Nagoya University Graduate School of Medicine. FUNDING: This work was financially supported by the Strategic Professional Development
Program for Young Researchers (MEXT) and the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (Grant Numbers 17K11230 and 22K09638). AUTHOR INFORMATION AUTHORS
AND AFFILIATIONS * Department of Obstetrics and Gynecology, Nagoya University Graduate School of Medicine, Nagoya, Aichi, 466‑8550, Japan Yukako Iitani, Kenji Imai, Kazuya Fuma, Takafumi
Ushida, Sho Tano, Kosuke Yoshida, Akira Yokoi, Hiroaki Kajiyama & Tomomi Kotani * Laboratory of Bell Research Center‑Department of Obstetrics and Gynecology Collaborative Research,
Nagoya University Graduate School of Medicine, Nagoya, Aichi, 466‑8550, Japan Rika Miki * Center for Maternal-Neonatal Care, Nagoya University Hospital, Nagoya, Aichi, 466‑8560, Japan
Takafumi Ushida & Tomomi Kotani * Nagoya University Institute for Advanced Research, Furo-cho, Chikusa-ku, Nagoya, 464-8603, Japan Kosuke Yoshida & Akira Yokoi Authors * Yukako
Iitani View author publications You can also search for this author inPubMed Google Scholar * Rika Miki View author publications You can also search for this author inPubMed Google Scholar *
Kenji Imai View author publications You can also search for this author inPubMed Google Scholar * Kazuya Fuma View author publications You can also search for this author inPubMed Google
Scholar * Takafumi Ushida View author publications You can also search for this author inPubMed Google Scholar * Sho Tano View author publications You can also search for this author
inPubMed Google Scholar * Kosuke Yoshida View author publications You can also search for this author inPubMed Google Scholar * Akira Yokoi View author publications You can also search for
this author inPubMed Google Scholar * Hiroaki Kajiyama View author publications You can also search for this author inPubMed Google Scholar * Tomomi Kotani View author publications You can
also search for this author inPubMed Google Scholar CONTRIBUTIONS Conception and design; Y.I., T.U., K.I. and T.K., Acquisition of data; Y.I., R.M., K.F., S.T. and T.K., Analysis and
interpretation of data; Y.I., T.U., K.I., R.M., K.Y., A.Y. and T.K., Drafting the article or revising it critically for important intellectual content; Y.I., T.U., K.I., A.Y., H.K., and
T.K.. Final approval of the version to be published; all authors. CORRESPONDING AUTHOR Correspondence to Tomomi Kotani. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE S1 RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a
publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing
agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Iitani, Y., Miki, R., Imai, K. _et al._ Interleukin-17A stimulation induces alterations in
Microglial microRNA expression profiles. _Pediatr Res_ 95, 167–173 (2024). https://doi.org/10.1038/s41390-023-02825-6 Download citation * Received: 01 April 2023 * Revised: 06 September 2023
* Accepted: 11 September 2023 * Published: 27 September 2023 * Issue Date: January 2024 * DOI: https://doi.org/10.1038/s41390-023-02825-6 SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative
Trending News
Page Not Found很抱歉,你所访问的页面已不存在了。如有疑问,请电邮[email protected]你仍然可选择浏览首页或以下栏目内容 :新闻生活娱乐财经体育视频播客新报业媒体有限公司版权所有(公司登记号:202120748H)...
Major Talbot Green shooting update as another murder arrest madeMajor Talbot Green shooting update as another murder arrest madeSix people have now been arrested on suspicion of murder...
'kiplinger' lists u. S. Bank as the best bank for retireesMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...
Chemical burn to the skin: a systematic review of first aid impacts on clinical outcomesBURNS Volume 48, Issue 7, November 2022, Pages 1527-1543 https://doi.org/10.1016/j.burns.2022.05.006Get rights and conte...
Executive travel : rental car fees might rise 10%-15%A 10% to 15% increase in car rental costs is expected to push 1995 business travel costs up 2%, according to a study by ...
Latests News
Interleukin-17a stimulation induces alterations in microglial microrna expression profilesABSTRACT BACKGROUND Increased maternal interleukin (IL)-17A and activated microglia are pivotal factors contributing to ...
Wired internet connections fall under new house guarantees in franceNEW COURT RULING CLARIFIES ISSUE FOR PROPERTY BUYERS If you buy a new-build property with wired internet, you are entitl...
Thank to you, chris, for sharing the story, the launch school experience and the whole process of…Thank to you, Chris, for sharing the story, the Launch School experience and the whole process of reflection that underl...
Eu urged to exempt drugs from bse rules - farmers weekly09 September 1997 EU URGED TO EXEMPT DRUGS FROM BSE RULES EUROPEAN Union scientists have advised that pharmaceuticals an...
Psychology doctoral internship program in health service psychology | veterans affairsThe psychology doctoral internship at VA Black Hills Health Care System is fully accredited by the Commission on Accredi...