Cooperative structure of the heterotrimeric pre-mrna retention and splicing complex
Cooperative structure of the heterotrimeric pre-mrna retention and splicing complex"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
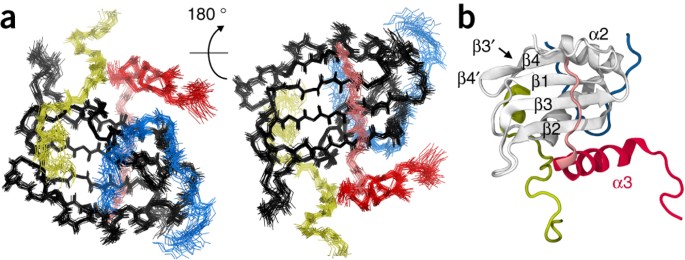
ABSTRACT The precursor mRNA (pre-mRNA) retention and splicing (RES) complex is a spliceosomal complex that is present in yeast and humans and is important for RNA splicing and retention of
unspliced pre-mRNA. Here, we present the solution NMR structure of the RES core complex from _Saccharomyces cerevisiae_. Complex formation leads to an intricate folding of three
components—Snu17p, Bud13p and Pml1p—that stabilizes the RNA-recognition motif (RRM) fold of Snu17p and increases binding affinity in tertiary interactions between the components by more than
100-fold compared to that in binary interactions. RES interacts with pre-mRNA within the spliceosome, and through the assembly of the RES core complex RNA binding efficiency is increased.
The three-dimensional structure of the RES core complex highlights the importance of cooperative folding and binding in the functional organization of the spliceosome. Access through your
institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print
issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to
local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS MECHANISM FOR THE INITIATION OF SPLICEOSOME DISASSEMBLY Article 26 June 2024 RNA RECOGNITION BY NPL3P REVEALS U2 SNRNA-BINDING COMPATIBLE WITH A CHAPERONE ROLE DURING
SPLICING Article Open access 07 November 2023 STRUCTURAL BASIS OF HUMAN U5 SNRNP LATE BIOGENESIS AND RECYCLING Article 11 March 2024 ACCESSION CODES PRIMARY ACCESSIONS BIOLOGICAL MAGNETIC
RESONANCE DATA BANK * 19766 PROTEIN DATA BANK * 2MKC REFERENCED ACCESSIONS PROTEIN DATA BANK * 2FHO * 2PEH REFERENCES * Will, C.L. & Luhrmann, R. Spliceosome structure and function.
_Cold Spring Harb. Perspect. Biol._ 3, a3003707 (2011). Article CAS Google Scholar * Brow, D.A. Allosteric cascade of spliceosome activation. _Annu. Rev. Genet._ 36, 333–360 (2002).
Article CAS PubMed Google Scholar * Dziembowski, A. et al. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. _EMBO J._ 23, 4847–4856
(2004). Article CAS PubMed PubMed Central Google Scholar * Gottschalk, A., Bartels, C., Neubauer, G., Luhrmann, R. & Fabrizio, P. A novel yeast U2 snRNP protein, Snu17p, is required
for the first catalytic step of splicing and for progression of spliceosome assembly. _Mol. Cell. Biol._ 21, 3037–3046 (2001). Article CAS PubMed PubMed Central Google Scholar * Tuo,
S., Nakashima, K. & Pringle, J.R. Apparent defect in yeast bud-site selection due to a specific failure to splice the pre-mRNA of a regulator of cell-type-specific transcription. _PLoS
ONE_ 7, e47621 (2012). Article CAS PubMed PubMed Central Google Scholar * Scherrer, F.W. Jr. & Spingola, M. A subset of Mer1p-dependent introns requires Bud13p for splicing
activation and nuclear retention. _RNA_ 12, 1361–1372 (2006). Article CAS PubMed PubMed Central Google Scholar * Schmidlin, T. et al. Single-gene deletions that restore mating
competence to diploid yeast. _FEMS Yeast Res._ 8, 276–286 (2008). Article CAS PubMed Google Scholar * Zhou, Y., Chen, C. & Johansson, M.J. The pre-mRNA retention and splicing complex
controls tRNA maturation by promoting TAN1 expression. _Nucleic Acids Res._ 41, 5669–5678 (2013). Article CAS PubMed PubMed Central Google Scholar * Spingola, M., Armisen, J. &
Ares, M. Jr. Mer1p is a modular splicing factor whose function depends on the conserved U2 snRNP protein Snu17p. _Nucleic Acids Res._ 32, 1242–1250 (2004). Article CAS PubMed PubMed
Central Google Scholar * Hausmann, S. et al. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small
nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways. _J. Biol. Chem._ 283, 31706–31718 (2008). Article CAS PubMed PubMed Central Google Scholar *
Wang, Q. & Rymond, B.C. Rds3p is required for stable U2 U2 snRNP recruitment to the splicing apparatus. _Mol. Cell. Biol._ 23, 7339–7349 (2003). Article CAS PubMed PubMed Central
Google Scholar * Wang, Q., He, J., Lynn, B. & Rymond, B.C. Interactions of the yeast SF3b splicing factor. _Mol. Cell. Biol._ 25, 10745–10754 (2005). Article CAS PubMed PubMed
Central Google Scholar * Brooks, M.A. et al. Structure of the yeast Pml1 splicing factor and its integration into the RES complex. _Nucleic Acids Res._ 37, 129–143 (2009). Article CAS
PubMed Google Scholar * Jiang, M. et al. Genome-wide analysis of developmental and sex-regulated gene expression profiles in _Caenorhabditis elegans_. _Proc. Natl. Acad. Sci. USA_ 98,
218–223 (2001). Article CAS PubMed Google Scholar * Trowitzsch, S., Weber, G., Luhrmann, R. & Wahl, M.C. An unusual RNA recognition motif acts as a scaffold for multiple proteins in
the pre-mRNA retention and splicing complex. _J. Biol. Chem._ 283, 32317–32327 (2008). Article CAS PubMed Google Scholar * Collinet, B. et al. Strategies for the structural analysis of
multi-protein complexes: lessons from the 3D-Repertoire project. _J. Struct. Biol._ 175, 147–158 (2011). Article CAS PubMed Google Scholar * Trowitzsch, S., Weber, G., Luhrmann, R. &
Wahl, M.C. Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex. _J. Mol. Biol._ 385, 531–541 (2009). Article CAS PubMed Google Scholar *
Dinkel, H. et al. ELM: the database of eukaryotic linear motifs. _Nucleic Acids Res._ 40, D242–D251 (2012). Article CAS PubMed Google Scholar * Korneta, I. & Bujnicki, J.M. Intrinsic
disorder in the human spliceosomal proteome. _PLoS Comput. Biol._ 8, e1002641 (2012). Article CAS PubMed PubMed Central Google Scholar * Kielkopf, C.L., Rodionova, N.A., Green, M.R.
& Burley, S.K. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. _Cell_ 106, 595–605 (2001). Article CAS PubMed Google Scholar *
Corsini, L. et al. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. _Nat. Struct. Mol. Biol._ 14, 620–629 (2007). Article CAS PubMed Google
Scholar * Selenko, P. et al. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. _Mol. Cell_ 11, 965–976 (2003). Article CAS PubMed Google
Scholar * Thickman, K.R., Swenson, M.C., Kabogo, J.M., Gryczynski, Z. & Kielkopf, C.L. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of
protein-protein interactions among pre-mRNA splicing factors. _J. Mol. Biol._ 356, 664–683 (2006). Article CAS PubMed Google Scholar * Shen, Y. & Bax, A. Protein backbone and
sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. _J. Biomol. NMR_ 56, 227–241 (2013). Article CAS PubMed PubMed Central Google Scholar *
Adam, S.A., Nakagawa, T., Swanson, M.S., Woodruff, T.K. & Dreyfuss, G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein
consensus sequence. _Mol. Cell. Biol._ 6, 2932–2943 (1986). Article CAS PubMed PubMed Central Google Scholar * Fabrizio, P. et al. The evolutionarily conserved core design of the
catalytic activation step of the yeast spliceosome. _Mol. Cell_ 36, 593–608 (2009). Article CAS PubMed Google Scholar * Ohrt, T. et al. Prp2-mediated protein rearrangements at the
catalytic core of the spliceosome as revealed by dcFCCS. _RNA_ 18, 1244–1256 (2012). Article CAS PubMed PubMed Central Google Scholar * Warkocki, Z. et al. Reconstitution of both steps
of _Saccharomyces cerevisiae_ splicing with purified spliceosomal components. _Nat. Struct. Mol. Biol._ 16, 1237–1243 (2009). Article CAS PubMed Google Scholar * Gozani, O., Feld, R.
& Reed, R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. _Genes Dev._
10, 233–243 (1996). Article CAS PubMed Google Scholar * McPheeters, D.S. & Muhlenkamp, P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region
during pre-mRNA splicing in yeast. _Mol. Cell. Biol._ 23, 4174–4186 (2003). Article CAS PubMed PubMed Central Google Scholar * Will, C.L. et al. A novel U2 and U11/U12 snRNP protein
that associates with the pre-mRNA branch site. _EMBO J._ 20, 4536–4546 (2001). Article CAS PubMed PubMed Central Google Scholar * Schellenberg, M.J. et al. Crystal structure of a core
spliceosomal protein interface. _Proc. Natl. Acad. Sci. USA_ 103, 1266–1271 (2006). Article CAS PubMed PubMed Central Google Scholar * Kuwasako, K. et al. Complex assembly mechanism and
an RNA-binding mode of the human p14–SF3b155 spliceosomal protein complex identified by NMR solution structure and functional analyses. _Proteins_ 71, 1617–1636 (2008). Article CAS PubMed
Google Scholar * Martin-Tumasz, S., Richie, A.C., Clos, L.J. II, Brow, D.A. & Butcher, S.E. A novel occluded RNA recognition motif in Prp24 unwinds the U6 RNA internal stem loop.
_Nucleic Acids Res._ 39, 7837–7847 (2011). Article CAS PubMed PubMed Central Google Scholar * Netter, C., Weber, G., Benecke, H. & Wahl, M.C. Functional stabilization of an RNA
recognition motif by a noncanonical N-terminal expansion. _RNA_ 15, 1305–1313 (2009). Article CAS PubMed PubMed Central Google Scholar * Avis, J.M. et al. Solution structure of the
N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. _J. Mol. Biol._ 257, 398–411 (1996). Article CAS PubMed Google Scholar *
Golovanov, A.P., Hautbergue, G.M., Tintaru, A.M., Lian, L.Y. & Wilson, S.A. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export
factors and RNA. _RNA_ 12, 1933–1948 (2006). Article CAS PubMed PubMed Central Google Scholar * Cléry, A. et al. Molecular basis of purine-rich RNA recognition by the human SR-like
protein Tra2-β1. _Nat. Struct. Mol. Biol._ 18, 443–450 (2011). Article PubMed CAS Google Scholar * Singh, M. et al. Structural basis for telomerase RNA recognition and RNP assembly by
the holoenzyme La family protein p65. _Mol. Cell_ 47, 16–26 (2012). Article CAS PubMed PubMed Central Google Scholar * Weber, G., Trowitzsch, S., Kastner, B., Luhrmann, R. & Wahl,
M.C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. _EMBO J._ 29, 4172–4184 (2010). Article CAS PubMed PubMed Central Google Scholar * Varani, L. et
al. The NMR structure of the 38 kDa U1A protein–PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. _Nat. Struct. Biol._ 7, 329–335
(2000). Article CAS PubMed Google Scholar * Williamson, J.R. Cooperativity in macromolecular assembly. _Nat. Chem. Biol._ 4, 458–465 (2008). Article CAS PubMed Google Scholar *
Berglund, J.A., Abovich, N. & Rosbash, M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. _Genes Dev._ 12, 858–867 (1998). Article CAS
PubMed PubMed Central Google Scholar * Corsini, L. et al. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. _J. Biol. Chem._ 284,
630–639 (2009). Article CAS PubMed Google Scholar * De Guzman, R.N., Goto, N.K., Dyson, H.J. & Wright, P.E. Structural basis for cooperative transcription factor binding to the CBP
coactivator. _J. Mol. Biol._ 355, 1005–1013 (2006). Article CAS PubMed Google Scholar * Brüschweiler, S. et al. Direct observation of the dynamic process underlying allosteric signal
transmission. _J. Am. Chem. Soc._ 131, 3063–3068 (2009). Article PubMed CAS Google Scholar * Hom, R.A. et al. Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
_J. Mol. Biol._ 400, 145–154 (2010). Article CAS PubMed PubMed Central Google Scholar * Cléry, A., Blatter, M. & Allain, F.H. RNA recognition motifs: boring? Not quite. _Curr.
Opin. Struct. Biol._ 18, 290–298 (2008). Article CAS PubMed Google Scholar * Daubner, G.M., Clery, A. & Allain, F.H. RRM-RNA recognition: NMR or crystallography.and new findings.
_Curr. Opin. Struct. Biol._ 23, 100–108 (2013). Article CAS PubMed Google Scholar * Daubner, G.M., Clery, A., Jayne, S., Stevenin, J. & Allain, F.H. A _syn_-_anti_ conformational
difference allows SRSF2 to recognize guanines and cytosines equally well. _EMBO J._ 31, 162–174 (2012). Article CAS PubMed Google Scholar * Oberstrass, F.C. et al. Structure of PTB bound
to RNA: specific binding and implications for splicing regulation. _Science_ 309, 2054–2057 (2005). Article CAS PubMed Google Scholar * Tsuda, K. et al. Structural basis for the dual
RNA-recognition modes of human Tra2-beta RRM. _Nucleic Acids Res._ 39, 1538–1553 (2011). Article CAS PubMed Google Scholar * Cléry, A. et al. Isolated pseudo-RNA-recognition motifs of SR
proteins can regulate splicing using a noncanonical mode of RNA recognition. _Proc. Natl. Acad. Sci. USA_ 110, E2802–E2811 (2013). Article PubMed PubMed Central Google Scholar *
Williams, S.G. & Hall, K.B. Binding affinity and cooperativity control U2B″/snRNA/U2A′ RNP formation. _Biochemistry_ 53, 3727–3737 (2014). Article CAS PubMed Google Scholar *
Williams, S.G. & Hall, K.B. Linkage and allostery in snRNP protein/RNA complexes. _Biochemistry_ 53, 3529–3539 (2014). Article CAS PubMed Google Scholar * Sattler, M., Schleucher, J.
& Griesinger, C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. _Prog. Nucl. Magn. Reson.
Spectrosc._ 34, 93–158 (1999). Article CAS Google Scholar * Yamazaki, T., Forman-Kay, J.D. & Kay, L.E. Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ɛ. chemical shifts
of aromatic residues in 13C-labeled proteins via scalar couplings. _J. Am. Chem. Soc._ 115, 11054–11055 (1993). Article CAS Google Scholar * Bax, A., Clore, G.M. & Gronenborn, A.M.
1H-1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. _J. Magn. Reson._ 88, 425–431 (1990).
CAS Google Scholar * Zwahlen, C. et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage λ N-peptide/boxB RNA complex. _J.
Am. Chem. Soc._ 119, 6711–6721 (1997). Article CAS Google Scholar * Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. _J. Biomol. NMR_ 6,
277–293 (1995). Article CAS PubMed Google Scholar * Vranken, W.F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. _Proteins_ 59, 687–696 (2005).
Article CAS PubMed Google Scholar * Yao, L., Ying, J. & Bax, A. Improved accuracy of 15N-1H scalar and residual dipolar couplings from gradient-enhanced IPAP-HSQC experiments on
protonated proteins. _J. Biomol. NMR_ 43, 161–170 (2009). Article CAS PubMed PubMed Central Google Scholar * Permi, P., Heikkinen, S., Kilpelainen, I. & Annila, A. Measurement of
1JNC′ and 2JHNC′ couplings from spin-state-selective two-dimensional correlation spectrum. _J. Magn. Reson._ 140, 32–40 (1999). Article CAS PubMed Google Scholar * Zweckstetter, M. NMR:
prediction of molecular alignment from structure using the PALES software. _Nat. Protoc._ 3, 679–690 (2008). Article CAS PubMed Google Scholar * Pagano, K. et al. Direct and allosteric
inhibition of the FGF2/HSPGs/FGFR1 ternary complex formation by an antiangiogenic, thrombospondin-1-mimic small molecule. _PLoS ONE_ 7, e36990 (2012). Article CAS PubMed PubMed Central
Google Scholar * Schanda, P., Kupce, E. & Brutscher, B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. _J.
Biomol. NMR_ 33, 199–211 (2005). Article CAS PubMed Google Scholar * Güntert, P., Mumenthaler, C. & Wuthrich, K. Torsion angle dynamics for NMR structure calculation with the new
program DYANA. _J. Mol. Biol._ 273, 283–298 (1997). Article PubMed Google Scholar * Nederveen, A.J. et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using
restraints from the BioMagResBank. _Proteins_ 59, 662–672 (2005). Article CAS PubMed Google Scholar * Schwieters, C.D., Kuszewski, J.J., Tjandra, N. & Clore, G.M. The Xplor-NIH NMR
molecular structure determination package. _J. Magn. Reson._ 160, 65–73 (2003). Article CAS PubMed Google Scholar * Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular
dynamics. _J. Mol. Graph._ 14, 33–38 (1996). Article CAS PubMed Google Scholar * Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems:
application to microtubules and the ribosome. _Proc. Natl. Acad. Sci. USA_ 98, 10037–10041 (2001). Article CAS PubMed PubMed Central Google Scholar * Yean, S.L. & Lin, R.J. U4 small
nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. _Mol. Cell. Biol._ 11, 5571–5577 (1991). Article CAS PubMed PubMed Central
Google Scholar * Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. _Methods_ 24, 218–229 (2001). Article CAS PubMed
Google Scholar * Moore, M.J. & Sharp, P.A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. _Science_ 256, 992–997 (1992). Article CAS PubMed
Google Scholar * Silverman, S.K. & Baum, D.A. Use of deoxyribozymes in RNA research. _Methods Enzymol._ 469, 95–117 (2009). Article CAS PubMed PubMed Central Google Scholar *
Urlaub, H., Hartmuth, K., Kostka, S., Grelle, G. & Luhrmann, R. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear
ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6.U5 snRNPs. _J. Biol. Chem._ 275, 41458–41468 (2000). Article CAS PubMed Google Scholar Download
references ACKNOWLEDGEMENTS This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Collaborative Research Center 860 (project B2 to M.Z.) and the DFG Research Unit 806 (project
A6 to M.C.W. and R.L.). We thank P. Fabrizio and K. Hartmuth for helpful discussions, T. Wandersleben for help with protein preparation and K. Giller for the preparation of expression
constructs. AUTHOR INFORMATION Author notes * Simon Trowitzsch Present address: Present address: European Molecular Biology Laboratory, Grenoble Outstation, Grenoble, France, and Unit for
Virus Host-Cell Interactions, Université Grenoble Alpes–European Molecular Biology Laboratory–National Center for Scientific Research, Grenoble, France., AUTHORS AND AFFILIATIONS *
Department for NMR-based Structural Biology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany Piotr Wysoczański, ShengQi Xiang, Francesca Munari, Stefan Becker & Markus
Zweckstetter * Department of Cellular Biochemistry, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany Cornelius Schneider, Simon Trowitzsch & Reinhard Lührmann *
Laboratory of Structural Biochemistry, Freie Universität Berlin, Berlin, Germany Markus C Wahl * German Center for Neurodegenerative Diseases (DZNE), Göttingen, Germany Markus Zweckstetter *
Center for Nanoscale Microscopy and Molecular Physiology of the Brain, University Medical Center, Göttingen, Germany Markus Zweckstetter Authors * Piotr Wysoczański View author publications
You can also search for this author inPubMed Google Scholar * Cornelius Schneider View author publications You can also search for this author inPubMed Google Scholar * ShengQi Xiang View
author publications You can also search for this author inPubMed Google Scholar * Francesca Munari View author publications You can also search for this author inPubMed Google Scholar *
Simon Trowitzsch View author publications You can also search for this author inPubMed Google Scholar * Markus C Wahl View author publications You can also search for this author inPubMed
Google Scholar * Reinhard Lührmann View author publications You can also search for this author inPubMed Google Scholar * Stefan Becker View author publications You can also search for this
author inPubMed Google Scholar * Markus Zweckstetter View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS P.W. designed the project, conducted
protein preparation and ITC and NMR data acquisition and analysis and wrote the paper; C.S. performed immunoprecipitation of Snu17p from spliceosomal complexes; S.X. performed NMR
experiments; F.M. performed NMR data analysis; S.T. designed the project and conducted peptide arrays and protein preparation; M.C.W. designed and supervised the project and interpreted
data; R.L. designed and supervised the project; S.B. designed and supervised the project; M.Z. designed and supervised the project and wrote the paper. CORRESPONDING AUTHOR Correspondence to
Markus Zweckstetter. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. INTEGRATED SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE 1 COMPARISON OF
1H-15N HSQC OF CSNU17P IN VARIOUS COMPLEXES AND SECONDARY CHEMICAL SHIFT OF FREE CPML1P AND CBUD13P. (A) 1H, 15N HSQC of cSnu17p in cPml1p–cSnu17p dimer, cBud13p–cSnu17p dimer, and cRES. (B)
1H, 15N HSQC of cSnu17p in cPml1p–cSnu17p dimer and cSnu17p monomer. (C) 1H, 15N HSQC of cSnu17p in cPml1p–cSnu17p dimer and cBud13p–cSnu17p dimer. (D) 1H, 15N HSQC of cSnu17p in
cPml1p–cSnu17p dimer and cSnu17p monomer. (E) Secondary chemical shift (Δδ) of free cBud13p. (F) Secondary chemical shift (Δδ) of free cPml1p. . SUPPLEMENTARY FIGURE 2 VALIDATION OF THE CRES
STRUCTURE WITH RESIDUAL DIPOLAR COUPLINGS (RDCS). Plots of experimental vs back-calculated RDCs before (upper panel) and after (lower panel) structure refinement with RDCs. RDCs from the
C-terminal α-helix (117-126) backbone amides are indicated in red, those of cPml1p in yellow and cBud13p in blue. SUPPLEMENTARY FIGURE 3 1H-15N CHEMICAL-SHIFT PERTURBATION (CSP) OF
CSNU17P–CBUD13P OR CSNU17P–CPML1P WHEN TITRATED WITH CPML1P OR CBUD13P, RESPECTIVELY. (A) Plot of CSP imposed on cSnu17p–cBud13p dimer when titrated with cPml1p. (B) The aforementioned plot
mapped onto the structure of cRES with spheres colored as described above; T49 is indicated (C) Residues that are common between this CSP experiment (when cSnu17p-cBud13 dimer is titrated
with cPml1p) and when cRES is titrated with RNA (CUUCAUCUUUUUG) are labeled. (D) Plot of CSP imposed on cSnu17p–cPml1p dimer when titrated with cBud13p. (E–F) The aforementioned plot mapped
onto the structure of cRES with spheres colored as described above; T49 is indicated. Only 224 to 238 residues of cBud13p are shown for clarity. Significant CSPs were grouped and color-coded
into three categories according to: medium (light pink) if 2σ>CSP>1σ, strong (pink) if 3σ>CSP>2σ, very strong (red) if CSP>3σ, were σ is the standard deviation of the mean.
SUPPLEMENTARY FIGURE 4 ANALYSIS OF CSNU17P MONOMER, CSNU17P–CPML1P DIMER, CSNU17P–CBUD13P DIMER AND CRES DYNAMICS. (A) Plot of residue specific hydrogen-deuterium exchange half-life H-D1/2
for cRES (black), cSnu17p–cPml1p dimer (yellow), cSnu17p–cBud13p dimer (blue), cSnu17p monomer (red). (B) R2 (black) and R1ρ (grey) plots (left panel) for the aforementioned complexes. Rex
estimates derived from R2 and R1ρ difference (right panel). Positions marked with an asterisk correspond to the five most broadened peaks in C). (C) Normalized intensity vs 1H full width at
half maximum (FWHM) of 1H, 15N HSQC peaks for the aforementioned complexes and cSnu17p monomer. Intensity values are offset between each group by 1. Please note that both cRES and
cSnu17p–cBud13p dimer experience a decrease in the overall tumbling due to apparent increase in size related to peptide binding and/or the presence of C-terminal α-helix. This effect is
small in cSnu17p–cPml1p dimer since cPml1p is shorter and C-terminal α-helix is folded. (D) S2 order parameter derived from the chemical shift for the aforementioned complexes. SUPPLEMENTARY
FIGURE 5 RES-RNA INTERACTION. (A) 1H, 15N chemical shift perturbation (CSP) imposed on cSnu17p in cSnu17p–cPml1p dimer and in cSnu17p–cBud13p dimer upon titration with CUUCAUCUUUUUG RNA.
CSP mapped on the structure of cRES (left of each graph). Significant CSPs were grouped and color-coded into three categories according to: medium (light pink) if 2σ>CSP>1σ, strong
(pink) if 3σ>CSP>2σ, very strong (red) if CSP >3σ, were σ is the standard deviation of the mean for the CUUCAUCUUUUUG to cRES titration. Only 224 to 238 residues of cBud13p are
shown for clarity. (B) CSP binding curves derived from a representative set of residues experiencing high CSP and residue-averaged dissociation constants (_K_d) of CUUCAUUCUUUUUG and all
four members of the cRES assembly pathway. (C) Uncropped western blots from main text Fig. 5a. From left to right, Ponceau-stained membrane, peroxidase-anti-peroxidase detection of RES-TAP
and autoradiography. SUPPLEMENTARY FIGURE 6 1H-15N NORMALIZED CHEMICAL-SHIFT PERTURBATION (CSP) IMPOSED ON CSNU17P IN CRES UPON TITRATION WITH VARIOUS RNAS. (A) NCSP mapped on the structure
of cRES (left of each graph). Significant NCSPs were grouped and color-coded into three categories according to: medium (light pink) if 2σ>NCSP>1σ, strong (pink) if 3σ>NCSP>2σ,
very strong (red) if NCS >3σ, were σ is the standard deviation of the mean. Only 224 to 238 residues of cBud13p are shown for clarity. The given RNA sequence is indicated above each
graph. (B) CSP binding curves derived from a representative set of residues experiencing high CSP upon ACGAAUUAGA titration and average binding affinity (below). * value derived from CSP of
two residues. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–6 (PDF 2505 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Wysoczański, P., Schneider, C., Xiang, S. _et al._ Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex. _Nat Struct Mol Biol_ 21, 911–918 (2014).
https://doi.org/10.1038/nsmb.2889 Download citation * Received: 14 February 2014 * Accepted: 15 August 2014 * Published: 14 September 2014 * Issue Date: October 2014 * DOI:
https://doi.org/10.1038/nsmb.2889 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative
Trending News
Oat growers to gain from opening of new processing plant - farmers weeklyABSTRACT Powerful emissions from the centres of nearby galaxies may represent dead quasars. Access through your institut...
Location, location, location: trump has the best spot in american politicsHistory can weigh heavily on a filmmaker, and that is what happens with “Amelia,” a disappointing rendering of the remar...
How to be happy: 5 easy ways to increase your happinessMemorial Day Sale! Join AARP for just $11 per year with a 5-year membership Join now and get a FREE gift. Expires 6/4 G...
Patient mutations alter atrx targeting to pml nuclear bodiesABSTRACT ATRX is a SWI/SNF-like chromatin remodeling protein mutated in several X-linked mental retardation syndromes. G...
Rear viewing system for safe reversing - farmers weekly26 OCTOBER 2001 ------------------------- REAR VIEWING SYSTEM FOR SAFE REVERSING SPALDINGS has now launched a full colou...
Latests News
Cooperative structure of the heterotrimeric pre-mrna retention and splicing complexABSTRACT The precursor mRNA (pre-mRNA) retention and splicing (RES) complex is a spliceosomal complex that is present in...
Meet the team to enhance your knowledge and gain insightWhether you are interested in buying, selling, recruiting for or optimising the value of your dental practice or lab - D...
Page Not Found (404) | WFAE 90.7 - Charlotte's NPR News SourceWe're sorry; the page you're looking for cannot be found. Please use the search option at the top of the page to find wh...
What is headless cms- why is so popular?The “head” in “headless CMS” refers to the frontend. A headless content management system consists primarily of an API a...
Police seeking suspect in south boston aggravated assaultCrime THE INCIDENT OCCURRED LAST SUNDAY, JUNE 16, NEAR THE LINCOLN TAVERN AND RESTAURANT. By John Waller June 24, 2024 L...