Evolution of darwin’s finches and their beaks revealed by genome sequencing
Evolution of darwin’s finches and their beaks revealed by genome sequencing"
- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
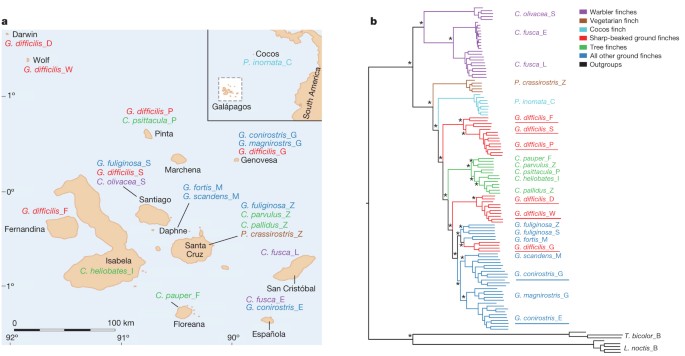
ABSTRACT Darwin’s finches, inhabiting the Galápagos archipelago and Cocos Island, constitute an iconic model for studies of speciation and adaptive evolution. Here we report the results of
whole-genome re-sequencing of 120 individuals representing all of the Darwin’s finch species and two close relatives. Phylogenetic analysis reveals important discrepancies with the
phenotype-based taxonomy. We find extensive evidence for interspecific gene flow throughout the radiation. Hybridization has given rise to species of mixed ancestry. A 240 kilobase haplotype
encompassing the _ALX1_ gene that encodes a transcription factor affecting craniofacial development is strongly associated with beak shape diversity across Darwin's finch species as
well as within the medium ground finch (_Geospiza fortis_), a species that has undergone rapid evolution of beak shape in response to environmental changes. The _ALX1_ haplotype has
contributed to diversification of beak shapes among the Darwin’s finches and, thereby, to an expanded utilization of food resources. Access through your institution Buy or subscribe This is
a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per
year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during
checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS THE BOWFIN GENOME
ILLUMINATES THE DEVELOPMENTAL EVOLUTION OF RAY-FINNED FISHES Article Open access 30 August 2021 GENOMIC SIGNATURES OF CONVERGENT SHIFTS TO PLUNGE-DIVING BEHAVIOR IN BIRDS Article Open access
24 October 2023 A CHROMOSOME-LEVEL GENOME OF THE STRIATED FROGFISH (_ANTENNARIUS STRIATUS_) Article Open access 21 June 2024 ACCESSION CODES PRIMARY ACCESSIONS GENBANK/EMBL/DDBJ * KM891730
SEQUENCE READ ARCHIVE * PRJNA263122 DATA DEPOSITS The Illumina reads have been submitted to the short reads archive (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA263122 and
the consensus sequence for the _G. fortis_ mtDNA has been submitted to GenBank under accession number KM891730. REFERENCES * Schluter, D. _The Ecology of Adaptive Radiation_ (Oxford Univ.
Press, 2000) Google Scholar * Seehausen, O. African cichlid fish: a model system in adaptive radiation research. _Proc. R. Soc. B_ 273, 1987–1998 (2006) Article PubMed PubMed Central
Google Scholar * Lack, D. _Darwin’s Finches_ (Cambridge Univ. Press, 1947) Google Scholar * Grant, P. R. _Ecology and Evolution of Darwin’s Finches_ (Princeton Univ. Press, 1999) Google
Scholar * Grant, P. R. & Grant, B. R. _How and Why Species Multiply. The Radiation of Darwin’s Finches_ (Princeton Univ. Press, 2008) Google Scholar * Petren, K., Grant, P. R., Grant,
B. R. & Keller, L. F. Comparative landscape genetics and the adaptive radiation of Darwin’s finches: the role of peripheral isolation. _Mol. Ecol._ 14, 2943–2957 (2005) Article CAS
PubMed Google Scholar * Ali, J. R. & Aitchison, J. C. Exploring the combined role of eustasy and oceanic island thermal subsidence in shaping biodiversity on the Galápagos. _J.
Biogeogr._ 41, 1227–1241 (2014) Article Google Scholar * Geist, D., Snell, H., Snell, H., Goddard, C. & Kurz, M. in _The Galápagos: A Natural Laboratory for the Earth Sciences_ (eds
Harpp K. S., Mittelstaedt E., d’Ozouville N., & Graham, D. ) 145–166 (American Geophysical Union, 2014) Google Scholar * Farrington, H. L., Lawson, L. P., Clark, C. M. & Petren, K.
The evolutionary history of Darwin’s finches: speciation, gene flow, and introgression in a fragmented landscape. _Evolution_ 68, 2932–2944 (2014) Article PubMed Google Scholar *
Abzhanov, A., Protas, M., Grant, B. R., Grant, P. R. & Tabin, C. J. Bmp4 and morphological variation of beaks in Darwin’s finches. _Science_ 305, 1462–1465 (2004) Article CAS ADS
PubMed Google Scholar * Abzhanov, A. et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. _Nature_ 442, 563–567 (2006) Article CAS ADS PubMed
Google Scholar * Mallarino, R. et al. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. _Proc. Natl Acad. Sci. USA_ 198, 4057–4062 (2011) Article ADS Google
Scholar * Burns, K. J. et al. Phylogenetics and diversification of tanagers (Passeriformes: Thraupidae), the largest radiation of Neotropical songbirds. _Mol. Phylogenet. Evol._ 75, 41–77
(2014) Article PubMed Google Scholar * Zhang, G., Parker, P., Li, B., Li, H. & Wang, J. The genome of Darwin’s finch (_Geospiza fortis_). _GigaScience_,
http://dx.doi.org/10.5524/100040 (3 August 2012) * Ellegren, H. The evolutionary genomics of birds. _Annu. Rev. Ecol. Evol. Syst._ 44, 239–259 (2013) Article Google Scholar * Balakrishnan,
C. N. & Edwards, S. V. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (_Taeniopygia guttata_). _Genetics_ 181,
645–660 (2009) Article PubMed PubMed Central Google Scholar * Swarth, H. S. The avifauna of the Galapagos Islands. _Occ. Pap. Calif. Acad. Sci._ 18, 1–299 (1931) Google Scholar * Lack,
D. The Galapagos finches (Geospizinae): a study in variation. _Occ. Pap. Calif. Acad. Sci._ 21, 1–159 (1945) Google Scholar * Durand, E. Y., Patterson, N., Reich, D. & Slatkin, M.
Testing for ancient admixture between closely related populations. _Mol. Biol. Evol._ 28, 2239–2252 (2011) Article CAS PubMed PubMed Central Google Scholar * Qvarnstrom, A. &
Bailey, R. I. Speciation through evolution of sex-linked genes. _Heredity_ 102, 4–15 (2009) Article CAS PubMed Google Scholar * Li, H. & Durbin, R. Inference of human population
history from individual whole-genome sequences. _Nature_ 475, 493–496 (2011) Article CAS PubMed PubMed Central Google Scholar * Rivera-Perez, J. A., Wakamiya, M. & Behringer, R. R.
Goosecoid acts cell autonomously in mesenchyme-derived tissues during craniofacial development. _Development_ 126, 3811–3821 (1999) CAS PubMed Google Scholar * Rowe, A., Richman, J. M.
& Brickell, P. M. Retinoic acid treatment alters the distribution of retinoic acid receptor-β transcripts in the embryonic chick face. _Development_ 111, 1007–1016 (1991) CAS PubMed
Google Scholar * Uz, E. et al. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia.
_Am. J. Hum. Genet._ 86, 789–796 (2010) Article CAS PubMed PubMed Central Google Scholar * Dee, C. T., Szymoniuk, C. R., Mills, P. E. D. & Takahashi, T. Defective neural crest
migration revealed by a zebrafish model of Alx1-related frontonasal dysplasia. _Hum. Mol. Genet._ 22, 239–251 (2013) Article CAS PubMed Google Scholar * Brugmann, S. A. et al.
Comparative gene expression analysis of avian embryonic facial structures reveals new candidates for human craniofacial disorders. _Hum. Mol. Genet._ 19, 920–930 (2010) Article CAS PubMed
Google Scholar * Sommer, P., Napier, H. R., Hogan, B. L. & Kidson, S. H. Identification of Tgfβ1i4 as a downstream target of Foxc1. _Dev. Growth Differ._ 48, 297–308 (2006) Article
CAS PubMed Google Scholar * Wang, J. et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. _Nucleic Acids Res._ 41,
D171–D176 (2013) Article CAS PubMed Google Scholar * Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT
algorithm. _Nature Protocols_ 4, 1073–1081 (2009) Article CAS PubMed Google Scholar * Grant, P. R. & Grant, B. R. _40 Years of Evolution. Darwin’s Finches on Daphne Major Island_
(Princeton Univ. Press, 2014) Book Google Scholar * Boag, P. T. Growth and allometry of external morphology in Darwin’s finches (_Geospiza_) on Isla Daphne Major, Galápagos. _J. Zool._
204, 413–441 (1984) Article Google Scholar * The Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. _Nature_ 487, 94–98 (2012)
Article ADS CAS PubMed Central Google Scholar * Andersson, L. Molecular consequences of animal breeding. _Curr. Opin. Genet. Dev._ 23, 295–301 (2013) Article CAS PubMed Google
Scholar * Linnen, C. R. et al. Adaptive evolution of multiple traits through multiple mutations at a single gene. _Science_ 339, 1312–1316 (2013) Article CAS ADS PubMed Google Scholar
* Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. _Genome Res._ 15, 1034–1050 (2005) Article CAS PubMed PubMed Central Google Scholar
* Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. _GigaScience_ 1, 18 (2012) Article PubMed PubMed Central Google Scholar * Li, H.
& Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. _Bioinformatics_ 25, 1754–1760 (2009) Article CAS PubMed PubMed Central Google Scholar * McKenna,
A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. _Genome Res._ 20, 1297–1303 (2010) Article CAS PubMed PubMed Central
Google Scholar * DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. _Nature Genet._ 43, 491–498 (2011) Article CAS PubMed
Google Scholar * Van der Auwera, G. A. et al. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. _Curr. Protoc. Bioinform._ 43,
11.10.1–11.10.33 (2002) Google Scholar * Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of
localized haplotype clustering. _Am. J. Hum. Genet._ 81, 1084–1097 (2007) Article CAS PubMed PubMed Central Google Scholar * Purcell, S. et al. PLINK: a tool set for whole-genome
association and population-based linkage analyses. _Am. J. Hum. Genet._ 81, 559–575 (2007) Article CAS PubMed PubMed Central Google Scholar * Green, R. E. et al. A draft sequence of the
Neandertal genome. _Science_ 328, 710–722 (2010) Article CAS ADS PubMed PubMed Central Google Scholar * Holm, S. A simple sequentially rejective multiple test procedure. _Scand. J.
Stat._ 6, 65–70 (1979) MathSciNet MATH Google Scholar * Rands, C. et al. Insights into the evolution of Darwin’s finches from comparative analysis of the _Geospiza magnirostris_ genome
sequence. _BMC Genomics_ 14, 95 (2013) Article CAS PubMed PubMed Central Google Scholar * Weir, J. T. & Schluter, D. Calibrating the avian molecular clock. _Mol. Ecol._ 17,
2321–2328 (2008) Article CAS PubMed Google Scholar * Watterson, G. A. On the number of segregating sites in genetical models without recombination. _Theor. Popul. Biol._ 7, 256–276
(1975) Article CAS MathSciNet PubMed MATH Google Scholar * Grant, B. R. & Grant, P. R. Demography and the genetically effective sizes of two populations of Darwin’s finches.
_Ecology_ 73, 766–784 (1992) Article Google Scholar * Nei, M. in _Molecular Evolutionary Genetics_ 276–279 (Columbia Univ. Press, 1987) Book Google Scholar * Danecek, P. et al. The
variant call format and VCFtools. _Bioinformatics_ 27, 2156–2158 (2011) Article CAS PubMed PubMed Central Google Scholar * Vilella, A. J. et al. EnsemblCompara GeneTrees: complete,
duplication-aware phylogenetic trees in vertebrates. _Genome Res._ 19, 327–335 (2009) Article CAS PubMed PubMed Central Google Scholar * Edgar, R. C. MUSCLE: multiple sequence alignment
with high accuracy and high throughput. _Nucleic Acids Res._ 32, 1792–1797 (2004) Article CAS PubMed PubMed Central Google Scholar * Waterhouse, A. M., Procter, J. B., Martin, D. M.
A., Clamp, M. & Barton, G. J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. _Bioinformatics_ 25, 1189–1191 (2009) Article CAS PubMed PubMed Central
Google Scholar * Jones, P. et al. InterProScan 5: genome-scale protein function classification. _Bioinformatics_ 30, 1236–1240 (2014) Article CAS PubMed PubMed Central Google Scholar *
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of _Drosophila melanogaster_ strain _w__1118_;
_iso-2_; _iso-3_. _Fly (Austin)_ 6, 80–92 (2012) Article CAS Google Scholar * Grant, B. R., Grant, P. R. & Petren, K. The allopatric phase of speciation: the sharp-beaked ground finch
(_Geospiza difficilis_) on the Galápagos islands. _Biol. J. Linn. Soc._ 69, 287–317 (2000) Article Google Scholar * Grant, P. R., Abbott, I., Schluter, D., Curry, R. L. & Abbott, L.
K. Variation in the size and shape of Darwin’s finches. _Biol. J. Linn. Soc._ 25, 1–39 (1985) Article Google Scholar * Schluter, D. & Grant, P. R. Ecological correlates of
morphological evolution in a Darwin’s finch, _Geospiza difficilis_. _Evolution_ 38, 856–869 (1984) Article PubMed Google Scholar * Rabosky, D. Diversity-dependence, ecological speciation,
and the role of competition in macroevolution. _Ann. Rev. Evol. Ecol. Syst._ 44, 481–502 (2013) Article Google Scholar Download references ACKNOWLEDGEMENTS The National Science Foundation
(USA) funded the collection of material under permits from the Galápagos and Costa Rica National Parks Services, and in accordance with protocols of Princeton University’s Animal Welfare
Committee. The map and images of finch heads are reproduced with permission from Princeton University Press. The project was supported by the Knut and Alice Wallenberg Foundation. Sequencing
was performed by the SNP&SEQ Technology Platform, supported by Uppsala University and Hospital, SciLifeLab and Swedish Research Council (80576801 and 70374401). Computer resources were
supplied by UPPMAX. AUTHOR INFORMATION Author notes * Sangeet Lamichhaney and Jonas Berglund: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Medical
Biochemistry and Microbiology, Uppsala University, SE-751 23 Uppsala, Sweden, Sangeet Lamichhaney, Jonas Berglund, Markus Sällman Almén, Manfred Grabherr, Alvaro Martinez-Barrio, Marta
Promerová, Carl-Johan Rubin, Chao Wang, Neda Zamani, Matthew T. Webster & Leif Andersson * Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences,
SE-75007 Uppsala, Sweden, Khurram Maqbool & Leif Andersson * Department of Plant Physiology, Umeå University, SE-901 87 Umeå, Sweden, Neda Zamani * Department of Ecology and Evolutionary
Biology, Princeton University, Princeton, 08544, New Jersey, USA B. Rosemary Grant & Peter R. Grant * Department of Veterinary Integrative Biosciences, Texas A&M University, College
Station, 77843-4458, Texas, USA Leif Andersson Authors * Sangeet Lamichhaney View author publications You can also search for this author inPubMed Google Scholar * Jonas Berglund View
author publications You can also search for this author inPubMed Google Scholar * Markus Sällman Almén View author publications You can also search for this author inPubMed Google Scholar *
Khurram Maqbool View author publications You can also search for this author inPubMed Google Scholar * Manfred Grabherr View author publications You can also search for this author inPubMed
Google Scholar * Alvaro Martinez-Barrio View author publications You can also search for this author inPubMed Google Scholar * Marta Promerová View author publications You can also search
for this author inPubMed Google Scholar * Carl-Johan Rubin View author publications You can also search for this author inPubMed Google Scholar * Chao Wang View author publications You can
also search for this author inPubMed Google Scholar * Neda Zamani View author publications You can also search for this author inPubMed Google Scholar * B. Rosemary Grant View author
publications You can also search for this author inPubMed Google Scholar * Peter R. Grant View author publications You can also search for this author inPubMed Google Scholar * Matthew T.
Webster View author publications You can also search for this author inPubMed Google Scholar * Leif Andersson View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS P.R.G. and B.R.G. collected the material. L.A., P.R.G. and B.R.G. conceived the study. L.A. and M.T.W. led the bioinformatic analysis of data. S.L. and J.B. performed
the bioinformatic analysis with contributions from M.S.A., K.M., M.G., A.M.-B., C.-J.R. and N.Z. M.P. and C.W. performed experimental work. L.A., S.L., J.B., B.R.G., P.R.G. and M.T.W. wrote
the paper with input from the other authors. All authors approved the manuscript before submission. CORRESPONDING AUTHOR Correspondence to Leif Andersson. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing financial interests. EXTENDED DATA FIGURES AND TABLES EXTENDED DATA FIGURE 1 READ DEPTH. Average read depth in all 120 samples of Darwin’s finches
and outgroup species. EXTENDED DATA FIGURE 2 GENETIC DIVERSITY AMONG DARWIN’S FINCHES. Heat map illustrating the proportion of shared and fixed polymorphisms among Darwin’s finches and
outgroup species. EXTENDED DATA FIGURE 3 NETWORK TREE FOR THE DARWIN’S FINCHES ON THE BASIS OF ALL AUTOSOMAL SITES. Taxa that showed deviations from classical taxonomy are underscored. Finch
heads are reproduced from ref. 5. _How and Why Species Multiply: The Radiation of Darwin's Finches_ by Peter R. Grant & B. Rosemary Grant. Copyright © 2008 Princeton University
Press. Reprinted by permission. EXTENDED DATA FIGURE 4 TAXONOMY AND RATE OF SPECIATION. A, Morphological variation among populations of ground finch (_Geospiza_) species, _scandens_,
_fuliginosa_ and three others, _acutirostris_, _difficilis_ and _septentrionalis_, that were formerly classified as a single species (_difficilis_). Data are from refs 56, 57, and from ref.
58 for weights and measures of _difficilis_ on Fernandina. B, Morphological variation among populations of _G. scandens_, _conirostris_, _propinqua_ and _magnirostris_ assessed by multiple
discriminant function analysis in JMP version 9. In a discriminant function analysis of the measured variables, all populations were correctly identified to species (−2 log likelihood _P_ =
0.02). Maximum discrimination was achieved by entering three variables in the sequence beak width, beak length and body size (weight or wing). Substituting beak depth for beak width gave the
same result. No other variable entered significantly. Data are from ref. 57, except for _scandens_ and _magnirostris_ data from ref. 30. C, Species accumulation on a log scale as a function
of time before the present, dating based on mtDNA. Species are expected to accumulate linearly according to a ‘birth–death’ process, eventually declining under a density- (diversity-)
dependent mechanism59. EXTENDED DATA FIGURE 5 PHYLOGENIES FOR MTDNA AND THE SEX CHROMOSOMES Z AND W. A, Tree based on mtDNA sequences. The dating of the nodes and their variances (in
thousands of years) is based on the cytochrome b sequences using the fossil-calibrated divergence rate 2.1% per million years for birds46. This tree based on the full mtDNA sequences shows
only minor differences compared with previously published trees based only on the cytochrome b sequence6,9. B, Maximum-likelihood trees based on all Z-linked sites; all nodes having full
local support on the basis of the Shimodaira–Hasegawa test are marked by asterisks. C, Tree based on W sequences, only females. Taxa that showed deviations from classical taxonomy are
underscored (applies to A–C). EXTENDED DATA FIGURE 6 ABBA–BABA ANALYSIS AND DEMOGRAPHIC HISTORY. A, ABBA–BABA analysis of _G. magnirostris_, _G. conirostris_ on Española and on Genovesa, and
with _L. noctis_ as outgroup. B, Comparison of _C. olivacea_, _C. fusca_, a pool of all non-warblers, and with _L. noctis_ as outgroup. The number of informative sites supporting the
different trees is indicated both as a percentage and as the actual number. The _D_ statistic and corresponding Holm–Bonferroni-corrected _P_ value are also given for testing the null
hypothesis of symmetry in genetic relationships. Finch heads are reproduced from ref. 5. C, PSMC analysis21 of all species except the _G. difficilis_ group. D, PSMC analysis of the _G.
difficilis_ group. EXTENDED DATA FIGURE 7 SEQUENCE CONSERVATION OF ALX1. Amino-acid alignment of the complete ALX1 sequence among different vertebrates. Amino-acid substitutions between
_ALX1_ alleles associated with blunt and pointed beaks are highlighted. The homeobox domain is indicated. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION This file contains Supplementary
Text and References. (PDF 275 kb) SUPPLEMENTARY TABLE 1 This file contains read depth in males and females for the identification of scaffolds from chromosome Z and W. Part a shows read
depth in males and females for scaffolds assigned to the Z chromosome. Part b shows read depth in males and females for scaffolds assigned to the W chromosome. (XLSX 79 kb) SUPPLEMENTARY
TABLE 2 This file contains details from ABBA-BABA analyses of Darwin’s finch populations. _P_-values are two-sided Holm-Bonferroni corrected. (XLSX 59 kb) POWERPOINT SLIDES POWERPOINT SLIDE
FOR FIG. 1 POWERPOINT SLIDE FOR FIG. 2 POWERPOINT SLIDE FOR FIG. 3 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lamichhaney, S., Berglund, J., Almén,
M. _et al._ Evolution of Darwin’s finches and their beaks revealed by genome sequencing. _Nature_ 518, 371–375 (2015). https://doi.org/10.1038/nature14181 Download citation * Received: 09
October 2014 * Accepted: 31 December 2014 * Published: 11 February 2015 * Issue Date: 19 February 2015 * DOI: https://doi.org/10.1038/nature14181 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative
Trending News
This Morning viewers ask 'what was that' as Holly Willoughby and Phillip Schofield 'replaced'This Morning viewers ask 'what was that' as Holly Willoughby and Phillip Schofield 'replaced'All eyes have been on the d...
Susan calman health: her mental health is no laughing matterAppearing on BBC Radio 4's The News Quiz and I'm Sorry I Haven't a Clue, Susan Calman is well-versed in w...
Avengers endgame, spider-man far from home trailers tease mcu future?If you watch the Avengers Endgame trailer and then Spider-Man Far From Home’s they’re certainly very different. We know ...
Fluid systems in chemical engineeringChemical Engineering Practice Edited By Herbert W. Cremer and Trefor Davies. Vol. 5: Fluid Systems I. Pp. vi + 695 + xxi...
Pollutants plucked from air with copperFortuitous catalyst discovery offers a new way to suck carbon dioxide from the atmosphere. Access through your instituti...
Latests News
Evolution of darwin’s finches and their beaks revealed by genome sequencingABSTRACT Darwin’s finches, inhabiting the Galápagos archipelago and Cocos Island, constitute an iconic model for studies...
Cypress finally gets elusive titleIt was just a matter of “Faith” as the Cypress College women’s soccer team ended a frustrating run of near-misses by win...
Agassi, hewitt: tale of agendasAndre Agassi hires a new coach and you could practically hear a sigh of relief from sponsors and tournament directors, a...
Lucy spraggan slams critics for reporting her post-boob job imagesBy RIANNE ADDO FOR MAILONLINE Published: 16:56 EDT, 11 January 2021 | Updated: 16:59 EDT, 11 January 2021 Lucy hit back ...
Supreme court upholds michigan's ban on affirmative actionThe Supreme Court on Tuesday ruled that citizens can legally nix affirmative action at the ballot box, thus upholding Mi...